Urea cycle regulation by mitochondrial sirtuin, SIRT5
Abstract
Mammalian sirtuins have diverse roles in aging, metabolism and disease. Recently we reported a new function for SIRT5 in urea cycle regulation. Our study uncovered that SIRT5 localized to mitochondria matrix and deacetylates carbamoyl phosphate synthetase 1 (CPS1), an enzyme which is the first and rate-limiting step of urea cycle. Deacetylation of CPS1 by SIRT5 resulted in activation of CPS1 enzymatic activity. Indeed, SIRT5-deficient mice failed to up-regulate CPS1 activity and showed hyper ammonemia during fasting. Similar effects are also observed on high protein diet or calorie restriction. These data indicate SIRT5 also has an emerging role in the metabolic adaptation to fasting, high protein diet and calorie restriction.
Sir2 is a NAD-dependent deacetylase and
promotes longevity in many organisms [1]. In mammals,
there are seven Sir2 homologues called sirtuins (SIRT1-7), which regulate
various biological functions in aging, metabolism and disease. Among of them,
SIRT3, SIRT4 and SIRT5 are believed to be localized in mitochondria [2]. SIRT3 is
the most characterized mitochondrial sirtuin. SIRT3 interacts with acetyl-CoA
synthetase 2 (ACS2) and deacetylates Lys-642 in vitro and in vivo.
Deacetylation of ACS2 by SIRT3 up-regulates the acetyl-CoA synthesis activity [3,4]. SIRT3
also deacetylates NDUFA9, one of the electron transport chain complex1
components to regulate ATP levels [5]. SIRT4
ADP-ribosylates glutamate dehydrogenase (GDH) and controls insulin secretion
in response to calorie restriction [6,7]. GDH is
also deacetylated by SIRT3, but its physiological significance is unknown [8]. These findings show that SIRT3 and SIRT4 directly
control the activity of metabolic enzymes in mitochondria and play an important
role in energy metabolism. However the
function of SIRT5 was unknown.
Recently we reported that
SIRT5 is localized in mitochondria matrix and regulates the urea cycle through
the deacetylation of CPS1 (Figure 1) [9]. By systemic
sub-fractionation of isolated mitochondria from mouse liver, we found SIRT5 was
also localized in the matrix fraction, as well as SIRT3 and SIRT4. This finding
corresponded with the fact that SIRT5 was cleaved in the N-terminus at a
typical consensus sequence recognized by mitochondria matrix peptidase. To
identify SIRT5-interacting proteins, we developed In vitro SIRT5-Flag
affinity purification, and found CPS1 as a SIRT5-binding protein. CPS1 is an
enzyme which mediates the first step of urea cycle. CPS1 is reported to be
acetylated at multiple lysine residues [10], however
the biological function of acetylation was unclear. Using an in vitro
deacetylation assay, we revealed that SIRT5 could deacetylate CPS1 in a
NAD-dependent manner and this deacetylation increased CPS1 enzymatic activity.
Indeed, SIRT5 deficient mice have ~30% lower CPS1 activity compared to wild
type mice. During fasting conditions, SIRT5 deficient mice failed to
up-regulate CPS1 activity and thereby resulted in hyper ammonemia. Similar results
were observed during calorie restriction or a high protein diet.
How does food limitation
activates the SIRT5? A previous study showed fasting induced the translocation
of NAD biosynthesis enzyme Nampt, to mitochondria and elevated the NAD levels
in mitochondria [11]. Although
we did not detect the translocation of Nampt to mitochondria in mouse liver, we
observed the increase of Nampt protein in the cytoplasm and the increase of NAD
level in the mitochondria. We hypothesize that the elevation of Nampt in
cytoplasm increase the NMN pool in the cytoplasm and NMN consequently
translocates into the mitochondria. In mitochondria, Nmnat3 converts NMN to
NAD, the next step in NAD biosynthesis, to activate SIRT5 (Figure 1). However
questions remain. For example, does Nampt translocation to mitochondria take
place in other tissues? How is NMN incorporated into mitochondria? In mitochondria, ~400
proteins have been shown to be acetylated [10]. However
little is known about their biological significance. As SIRT5 is expressed
ubiquitously in various tissues, SIRT5 probably has other substrates besides
CPS1. In fact, we also identified a few other SIRT5 interacting proteins using same strategy (our
unpublished data). Compared to deacetylation, little is known about the
mechanism of acetylation of mitochondrial proteins. Is acetylation in the
mitochondria enzymatic? If so, how many acetyltransferase are there, and what
is the specificity? More studies will be necessary to understand the biological
meaning of acetylation/deacetylation interplay in the mitochondria.
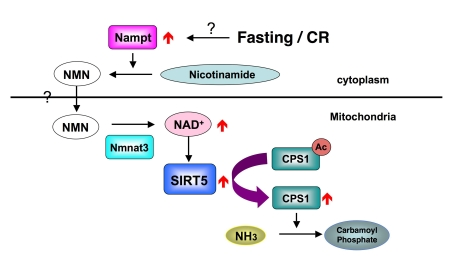
Figure 1. How SIRT5 is regulated during fasting and CR.
Interestingly, SIRT4
ADP-ribosylates GDH, which mediates the oxidative deamination step during
ammonia detoxification and represses GDH activity (Figure 2). Our findings show
that SIRT5 controls the subsequent step of the ammonia detoxification pathway.
Furthermore, OTC, an enzyme mediating the second step of the urea cycle in the
mitochondria matrix, was also recently reported to be regulated by reversible acetylation of Lysine-88 [12]. Deacetylation of OTC Lysine-88
increases the OTC enzymatic activity. In our study, no obvious change was
observed in OTC activity in SIRT3, SIRT4 and SIRT5 deficient mice under normal
conditions. However, a more detailed study under stressed condition, such as
fasting or calorie restriction may reveal the roles of mitochondrial sirtuins
in OTC deacetylation and the coordination of mitochondrial sirtuins in the
ammonia detoxification pathway.
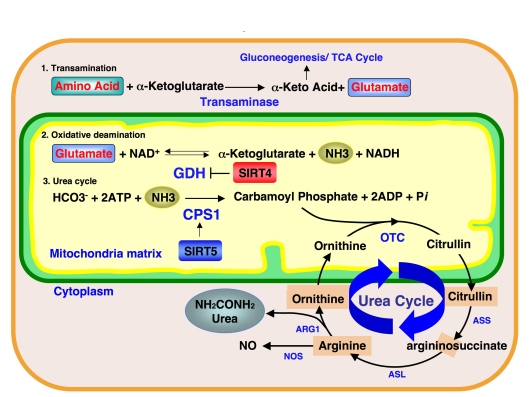
Figure 2. Ammonia detoxification pathway and mitochondrial sirtuins.
Our findings uncovered that, SIRT5 also has a pivotal
role in the metabolic adaptations during dietary shifts as well as other
sirtuins and co-factor NAD biosynthesis pathway is also the important regulator
of these processes. Drugs that activate SIRT5 may have therapeutic value to
treat hyper ammonemia.
Acknowledgments
This work was supported by a grant from Human Frontier
Science Program to T.N. and grants from the NIH and the Paul F. Glenn Foundation
to L.G.
Conflicts of Interest
L.G. is a consultant for Sirtris Pharmaceuticals.
References
-
1.
Guarente
L
Mitochondria--a nexus for aging, calorie restriction, and sirtuins.
Cell.
2008;
132:
171
-176.
[PubMed]
.
-
2.
Haigis
MC
and Guarente
LP.
Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction.
Genes Dev.
2006;
20:
2913
-2921.
[PubMed]
.
-
3.
Hallows
WC
, Lee
S
and Denu
JM.
Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases.
Proc Natl Acad Sci U S A.
2006;
103:
10230
-10235.
[PubMed]
.
-
4.
Schwer
B
, Bunkenborg
J
, Verdin
RO
, Andersen
JS
and Verdin
E.
Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2.
Proc Natl Acad Sci U S A.
2006;
103:
10224
-10229.
[PubMed]
.
-
5.
Ahn
BH
, Kim
HS
, Song
S
, Lee
IH
, Liu
J
, Vassilopoulos
A
, Deng
CX
and Finkel
T.
A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis.
Proc Natl Acad Sci U S A.
2008;
105:
14447
-14452.
[PubMed]
.
-
6.
Haigis
MC
, Mostoslavsky
R
, Haigis
KM
, Fahie
K
, Christodoulou
DC
, Murphy
AJ
, Valenzuela
DM
, Yancopoulos
GD
, Karow
M
, Blander
G
, Wolberger
C
, Prolla
TA
and Weindruch
R.
SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells.
Cell.
2006;
126:
941
-954.
[PubMed]
.
-
7.
Ahuja
N
, Schwer
B
, Carobbio
S
, Waltregny
D
, North
BJ
, Castronovo
V
, Maechler
P
and Verdin
E.
Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase.
J Biol Chem.
2007;
282:
33583
-33592.
[PubMed]
.
-
8.
Lombard
DB
, Alt
FW
, Cheng
HL
, Bunkenborg
J
, Streeper
RS
, Mostoslavsky
R
, Kim
J
, Yancopoulos
G
, Valenzuela
D
, Murphy
A
, Yang
Y
, Chen
Y
and Hirschey
MD.
Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation.
Mol Cell Biol.
2007;
27:
8807
-8814.
[PubMed]
.
-
9.
Nakagawa
T
, Lomb
DJ
, Haigis
MC
and Guarente
L.
SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle.
Cell.
2009;
137:
560
-570.
[PubMed]
.
-
10.
Kim
SC
, Sprung
R
, Chen
Y
, Xu
Y
, Ball
H
, Pei
J
, Cheng
T
, Kho
Y
, Xiao
H
, Xiao
L
, Grishin
NV
, White
M
and Yang
XJ.
Substrate and functional diversity of lysine acetylation revealed by a proteomics survey.
Mol Cell.
2006;
23:
607
-618.
[PubMed]
.
-
11.
Yang
H
, Yang
T
, Baur
JA
, Perez
E
, Matsui
T
, Carmona
JJ
, Lamming
DW
, Souza-Pinto
NC
, Bohr
VA
, Rosenzweig
A
, de Cabo
R
, Sauve
AA
and Sinclair
DA.
Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival.
Cell.
2007;
130:
1095
-1107.
[PubMed]
.
-
12.
Yu
W
, Lin
Y
, Yao
J
, Huang
W
, Lei
Q
, Xiong
Y
, Zhao
S
and Guan
KL.
Lysine 88 acetylation negatively regulates ornithine carbamoyltransferase activity in response to nutrient signals.
J Biol Chem.
2009;
284:
13669
-13675.
[PubMed]
.