Abstract
Gene expression patterns vary dramatically in a tissue-specific and age-dependent manner. RNA-binding proteins that regulate mRNA turnover and/or translation (TTR-RBPs) critically affect the subsets of expressed proteins. However, very little is known regarding the tissue- and age-dependent expression of TTR-RBPs in humans. Here, we use human tissue arrays containing a panel of organ biopsies from donors of different ages, to study the distribution and abundance of four TTR-RBPs: HuR, AUF1, TIA-1, and TTP. HuR and AUF1 were expressed with remarkably similar patterns. Both TTR-RBPs were present in high percentages of cells and displayed elevated intensities in many age groups and tissues, most notably in the gastrointestinal and reproductive systems; they were moderately expressed in the urinary and immune systems, and were almost undetectable in muscle and brain. TIA-1 was also abundant in many tissues and age groups; TIA-1 was expressed at high levels in the gastrointestinal, immune, urinary, and reproductive systems, and at low levels in brain and muscle. By contrast, TTP-expressing cells, as well as TTP signal intensities declined with advancing age, particularly in the immune, nervous, and muscular systems; however, TTP levels remained elevated in the gastrointestinal tract. The widespread abundance of HuR, AUF1, and TIA-1 throughout the body and in all age groups was in stark contrast with their declining levels in human diploid fibroblasts (HDFs) undergoing replicative senescence, a cultured-cell model of aging. Conversely, TTP levels increased in senescent HDFs, while TTP levels decreased with advancing age. Our studies provide a framework for the study of human TTR-RBP function in different tissues, throughout the human life span.
Introduction
Throughout the lifetime of an organism,
gene expression programs change dynamically. The specific subsets of proteins
expressed at each point in time allow cells to carry out long-term functions, such
as those needed during development and differentiation, and short-term adaptive
changes, including responses to acute environmental or hormonal stimuli. The
gene ex-pression patterns that characterize
each tissue at different developmental stages are strongly
regulated at the transcriptional level. Transcription factors (TFs) such as
FOXO (forkhead box), PPAR (peroxisome proliferator-activated receptor)γ, p53, C/EBP (CCAAT/enhanncer-binding protein), as well as by
chromatin remodeling factors such as MRG and HDACs have been implicated in
aging and age-related processes [1-6].
However, gene
expression patterns are also potently regulated by RNA-binding proteins (RBPs),
which control post-transcriptional processes such as
pre-mRNA splicing, and mRNA cytoplasmic export, turnover, storage, and
translation [7-10]. Unlike TFs, much less is known about the role of RBPs in
aging and age-related events. A subset of RBPs which function as t
ranslation
and t
urnover r
egulatory
(TTR) RBPs is of particular interest, since numerous genes implicated in
age-related processes encode mRNAs that are labile and/or subject to
translational control [11]. Examples of age-related proteins whose mRNAs are
targets of TTR-RBPs include p16INK4, p21CIP1, cyclins
(D1, E, A, B1, and H), cdk1 (cyclin-dependent kinase 1), CAK (cdk-activating
kinase), amyloid precursor protein (APP), endothelin-1, fibronectin,
interleukin (IL)-1, Cu,Zn- and Mn-superoxide dismutase (SOD), growth arrest- and
DNA damage-inducible (GADD)45, plasminogen activator inhibitor (PAI)-1 and
PAI-2, collagenase, granulocyte macrophage-colony-stimulating factor (GM-CSF)
and M-CSF, p53, bcl-2, p33ING1, c-fos, catalase, E2F-1,-2, DP-1,
elastin, thymidine kinase, insulin growth factor (IGF)-II, dihydrofolate
reductase, PCNA, ribonucleotide reductase, and histones (reviewed in [11]).
Here, we use arrays of human tissue biopsies to study the tissue distribution
of four major TTR-RBPs as a function of age: HuR (human antigen R), AUF1
(AU-binding factor 1, also called heterogenous ribonucleoprotein D or hnRNP D),
TIA (T-cell intracellular antigen)-1, and TTP (tristetraprolin).
HuR is the ubiquitously expressed member
of the embryonic lethal abnormal vision (ELAV)/Hu protein family, which also
comprises the primarily neuronal proteins HuB, HuC, and HuD [12]. Through its
RNA-recognition motifs (RRMs), HuR binds to numerous mRNAs bearing AU- and
U-rich sequences and stabilizes and/or modulates their translation [12-14]. Many
HuR target mRNAs encode proteins important for cell growth, proliferation, and
survival, as well as for the immune and stress responses [11,12,15-17].
Examples include mRNAs that encode cyclins (A, B1, E, D1), c-fos, c-myc,
vascular endothelial growth factor (VEGF), hypoxia-inducible factor-1α
(HIF-1α), prothymosin-α, cyclooxygenase (COX)-2, tumor necrosis
factor (TNF)-α, and several interleukins (reviewed in [11,12]).
AUF1 comprises four proteins
that arise from alternative splicing (p37, p40, p42, p45) and shuttle between
the nucleus and the cytoplasm [18,19]. AUF1 has also been implicated in
several distinct post-transcriptional processes. Originally found to promote
mRNA decay, as revealed in studies using a variety of cell systems [20-23], in
some instances AUF1 has been shown to enhance mRNA stability and to promote
translation [21,24-26]. All of the AUF1 isoforms contain two RRMs through
which they bind to a select group of mRNAs, including many mRNAs that encode
stress-response, immune, and proliferative proteins such as p21, cyclin D1,
myc, fos, GM-CSF, TNF-α, IL-3, parathyroid hormone (PTH), and GADD45α
[21-24,27,28].
TIA-1 and the TIA-1-related protein (TIAR)
are believed to play general roles as translational repressors in response to
environmental stress agents (heat, oxidants, hyperosmolarity, etc.) [29-33].
Such damaging factors trigger the aggregation of TIA-1 into discrete
cytoplasmic foci called stress granules (SGs), wherein mRNAs are thought to be
stored transiently and subsequently sorted into the translation and degradation
machineries. Many TIA-1/TIAR target mRNAs, often C-rich or U-rich [33,34],
are translationally repressed when they are associated with TIA-1/TIAR and
become translated upon dissociation from TIA-1 [27,33,35]. TIA-1 regulates
the translation of mRNAs encoding TNF-α, COX-2, and several other
transcripts bearing a TIA-1 motif [29,31,36].
The product of the ZFP-36 gene, TTP is
also known as TIS11, Nup475, and GOS24. TTP binds mRNAs through two tandem
CCCH zinc finger motifs and promotes their decay [37]. TTP target mRNAs
typically contain the AU-rich sequence UUAUUUAUU, although TTP also binds to
tandem repeats of the shorter sequence UUAU [38,39]. TTP is induced as an
immediate-early response gene in response to inflammatory mediators and growth
factors in many cell types, including T cells, macrophages, and fibroblasts
[40-43]. By destabilizing one of its target transcripts, TNF-α mRNA, TTP
reduced inflammation [40]. TTP also induced the decay of other mRNAs, such as
those encoding GM-CSF, COX-2, IL-3, IL-10, and interferon-γ [44-48].
We previously reported that cultured human
diploid fibroblasts (HDFs) undergoing replicative senescence showed reduced
levels of HuR, which contributed to the diminished expression of cyclin A,
cyclin B1, and c-fos in senescent HDFs [49], and also showed reduced AUF1
levels, which contributed to elevating p16 abundance, a senescence marker in this
cell system [50]. Fibroblasts explanted from donors of different ages, then
briefly expanded in culture, also showed a moderate reduction in HuR levels as
the age of the donor increased [49]. There is agreement that senescent HDFs
recapitulate some features of cells from the elderly and there is broad support
for the notion that cellular senescence constitutes a tumor suppressive
mechanism, particularly in young and middle-aged individuals. However, the
links between cellular senescence and aging or age-related processes are
considerably weaker.
A systematic study of TTR-RBPs in human tissues has not been performed
to-date. Yet many age-related genes are encoded by mRNAs which are labile
and/or subject to translational regulation. Therefore it was important to investigate the expression of TTR-RBPs, particularly HuR,
AUF1, TIA-1, and TTP, in a panel of human tissues spanning different ages. We
quantified both the percentages of TTR-RBP-positive cells and their relative
intensity as a function of individual donor age. This analysis provided a
wealth of information; salient among it was the finding that HuR, AUF1, and
TIA-1 remained highly expressed in many aging tissues, particularly in
gastrointestinal (GI) and reproductive organs, despite their reduced abundance
in replicative senescent HDFs. It was also interesting to discover that TTP
expression pattern was opposite to that of other TTR-RBPs, increasing with
replicative senescence and decreasing in many tissues with advancing age.
These findings reveal an important discordance between TTR-RBP levels during
replicative senescence and those present during in vivo aging, and
provide a valuable framework of tissue- and age-dependent TTR-RBP expression
for future in vivo analyses. Furthermore, they suggest that HuR, AUF1,
and TIA-1 likely play important roles in maintaining tissue homeostasis with
advancing age.
Results
HuR, AUF1, and TIA-1 levels decrease and TTP levels
increase during replicative senescence
To investigate the relative changes in
TTR-RBP levels occurring with replicative senescence and with increasing age,
we began by assessing TTR-RBP abundance in HDFs. Early-passage, proliferating
(‘young') WI-38 HDFs were cultured until they ceased cell division and became
senescent, as previously reported [51]. At the indicated population doublings
(pdls), protein lysates were prepared and the levels of HuR, AUF1, TIA-1, and
TTP were detected by Western blot analysis. As shown in Figure 1, HuR, AUF1,
and TIA-1 were most abundant in early-passage HDFs (pdl 21), declining
thereafter, as previously reported for HuR and AUF1 [49,50]. The levels of AUF1 and TIA-1 declined markedly by pdls 33 and 43,
becoming virtually undetectable by pdl 52, when cells were senescent, while the
decline in HuR levels was slower and less pronounced. Unexpectedly, the
expression pattern of TTP was just the opposite, displaying extremely low
levels in early-passage cells and increasing dramatically as WI-38 HDFs reached
senescence. The levels of GAPDH were measured to ensure equal loading.
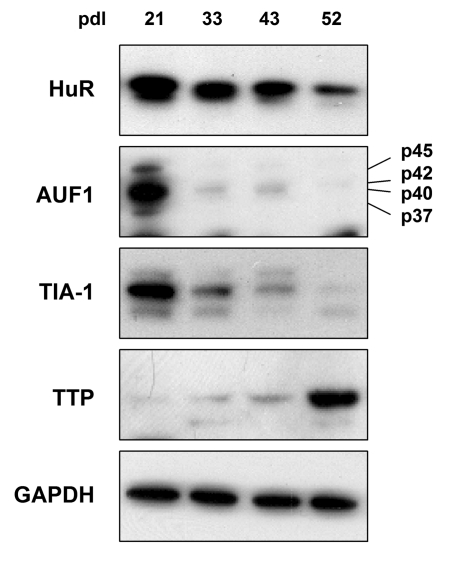
Figure 1. TTR-RBP expression in WI-38 human diploid fibroblasts (HDFs). WI-38 HDFs
were cultured for extended population doublings (pdls), until they reached
senescence at ~pdl 52. The abundance of TTR-RBPs HuR, AUF1 (all four
isoforms indicated), TIA-1, and TTP was assessed by Western blot analysis.
GAPDH signals were included as a loading control.
HuR levels remain elevated in numerous tissues with advancing age
In order to study the tissue- and age-dependent
expression patterns of TTR-RBPs, we obtained tissue arrays which contained a panel
of healthy tissue biopsies from human donors of different ages (fetal through
adult; Array II, BioChain Institute; FDA 35, Pantomics, Inc.). The tissue arrays
were probed with an anti-HuR antibody in order to visualize HuR signals in the
different tissues; the slides were then scanned and the digital images were
analyzed as explained in the Methods section. For the analysis, the donors'
ages were grouped as follows: fetal (F), young (Y, birth to 30
years of age), middle-aged (M, 30 to 60 years of age), or old (O,
over 60 years of age). The exact ages and tissue types of the biopsies
analyzed in both arrays are listed (Supplementary Table 1). The signals of
each spot on the array were measured in two ways: by counting the percentage area of positive cells (% HuR positive)
and by measuring the intensity of the signals in
the positive cells (Intensity). These values were calculated from the
digitized images using a color deconvolution algorithm to identify
diaminobenzidine (DAB, "brown") positivity in defined regions of interest (ROI)
[52]. The data were tabulated showing the number of samples analyzed in each age group in parenthesis, and the average
percentage values with the color scheme shown; in some cases, a tissue in a
given age group was not available in either of the arrays studied (n.a.).
Negative immunohistochemis-try signals are shown in the Supplementary Figure 1.
Table 1. Quantitation of HuR signals in human tissue microarrays.
Shown are the
percentages of positive area (‘% HuR positive') and the signal strength
(‘Intensity') in samples from a range of tissue types and age groups. When
multiple biopsies were quantified in a given tissue and age group, the
average value is shown and the number of tissues examined is indicated in
parenthesis. Values were calculated as explained in the Methods section.
As observed, HuR-positive cells were detected in
virtually all tissues and age groups (Table 1, left columns), but were very low
in neuronal and muscle tissues. The numbers of HuR-positive cells remained
relatively unchanged with increasing age in most tissues examined, increasing
with age only in the lung and in the gastrointestinal (GI) tract (small
intestine, colon, rectum). When HuR intensities were compared (Table 1, right
columns), there was little loss in HuR abundance with advancing age in most
tissues examined, declining only in the nervous system. Most tissues showed
little change in HuR levels across age groups (e.g., skeletal muscle, skin, and
reproductive and urinary systems), although an increase was observed again in
the lung. It is worth noting that strong HuR
sig- nals were seen throughout age groups in the GI,
reproductive, and urinary systems.
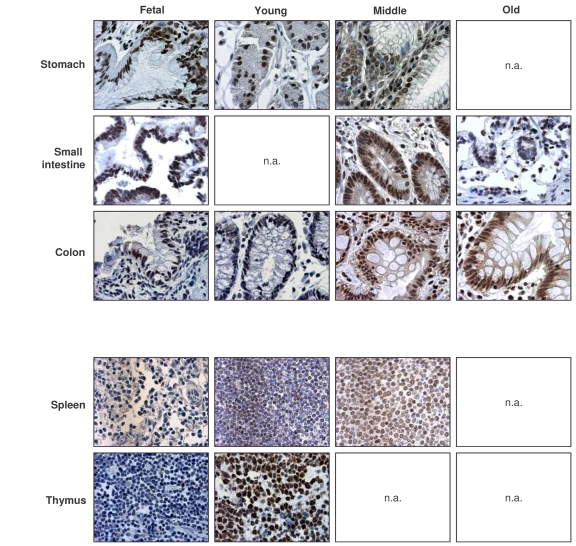
Figure 2. Immunohistochemical detection of HuR across tissue types and age groups.
Representative
HuR signals in photomicrographs taken from the indicated tissue sections
from human tissue arrays. Images are shown at ×200 magnification.
Representative photographs from the tissue array are
shown (Figure 2). Samples from the GI tract (stomach, small intestine, colon)
and the immune system (spleen, thymus) were selected, as the levels and
age-dependent changes in these tissues were particularly interesting for all
TTR-RBPs examined. In summary, HuR was ubiquitously expressed in a broad range
of human tissues and showed strong intensity despite advancing age. These
results contrasted with the loss of HuR expression seen in senescent HDFs ([49], Figure 1), and suggest that HuR remains
functionally important with advancing age.
Table 2. Quantitation of AUF1 signals in human tissue microarrays.
Shown are the
percentages of positive area (‘% AUF1 positive') and the signal strength
(‘Intensity') in samples from a range of tissue types and age groups. When
multiple biopsies were quantified in a given tissue and age group, the
average value is shown and the number of tissues examined is indicated in
parenthesis. Values were calculated as explained in the Methods section.
AUF1 expression is ubiquitous and overall abundant,
increasing with age in the immune system
The analysis of AUF1 in tissue arrays
was performed as described above for HuR. Interestingly, the relative
percentages of AUF1-expressing cells throughout the age groups, as well as the
relative intensities of AUF1 signals were rather similar to those seen for HuR
(compare Table 2 with Table 1); the tight correlation between AUF1 and HuR
signals was quantified (Supplementary Figure 2). A similar correlation
between AUF1 and HuR expression levels was noted by Lu and Schneider, who
compared their relative abundance in adult mouse tissues [53].
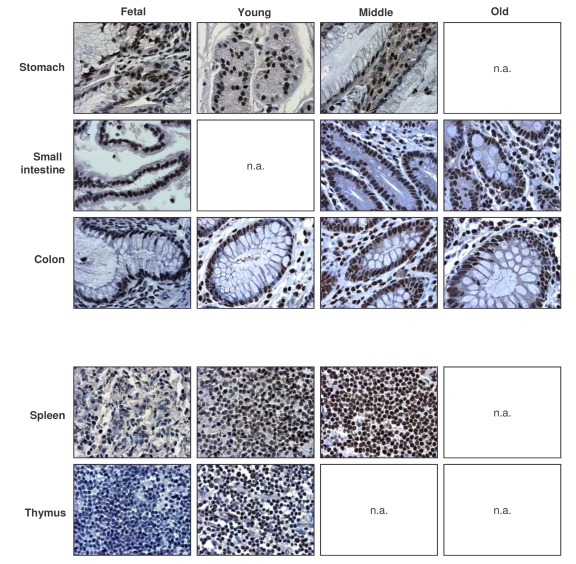
Figure 3. Immunohistochemical detection of AUF1 across tissue types and age groups.
Representative AUF1 signals in photomicrographs taken from the indicated
tissue sections from human tissue arrays. Images are shown at ×200
magnification.
AUF1-positive cells were detected
in all tissues examined, but were low in muscle, and high in the GI and immune
systems. AUF1 abundance increased with age in the immune system and was
overall high in the lung, GI tract, and urinary and reproductive systems.
Representative photographs of AUF1 expression in the GI and immune tissues are
shown in Figure 3. As seen with HuR, there was discordance between the steep
decline in AUF1 levels in senescent HDFs and the markedly elevated AUF1 levels
seen in tissues from elderly donors (Table 2). These findings support the
notion that AUF1 also plays a functional role in the tissues of elderly
individuals.
Table 3. Quantitation of TIA-1 signals in human tissue microarrays.
Shown are the
percentages of positive area (‘% TIA-1 positive') and the signal strength
(‘Intensity') in samples from a range of tissue types and age groups. When
multiple biopsies were quantified in a given tissue and age group, the
average value is shown and the number of tissues examined is indicated in
parenthesis. Values were calculated as explained in the Methods section.
Broad expression of TIA-1 across tissues and age groups
While TIA-1 also displayed a
ubiquitous distribution, TIA-1-positive cells showed a moderate decline in some
tissues of the GI and muscle systems (Table 3, left columns). Unlike HuR and
AUF1, the relative intensity of TIA-1 in several tissues declined with
advancing age, as seen in the endocrine, urinary, and muscle systems. Despite
a moderate decline in TIA-1 signals in the GI tract, its levels remained
relatively high here and were also elevated in all age groups in the
respiratory, immune, and reproductive systems (Table 3, right columns). Sample
photographs of TIA-1 signals in immune and GI specimens are shown in Figure 4.
Once again, TIA-1 followed a time-dependent pattern of expression in tissues
that was largely opposite to what was seen in cultured WI-38 HDFs advancing
towards senescence: highly expressed in vivo (Table 3, Figure 4),
progressive-ly lower until almost undetectable in vitro (Figure 1).
These observations suggest that TIA-1 may also be important for regulating gene expressionwith advancing age.
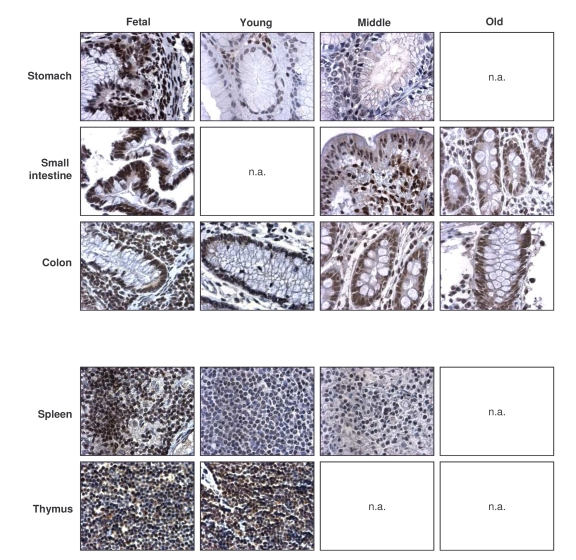
Figure 4. Immunohistochemical detection of TIA-1 across tissue types and age groups. Representative
TIA-1 signals in photomicrographs taken from the indicated tissue sections
from human tissue arrays. Images are shown at ×200 magnification.
General decline in TTP-expressing cells and TTP levels
with advancing age
Like TIA-1, the numbers of TTP-expressing cells were
highest in the fetal (F) group, generally decreasing in older groups (Table 4,
left columns). Exceptions to this pattern were the GI and endocrine systems,
where TTP-positive cell numbers remained constant across age groups, and the
reproductive tissues, where TTP-positive cells increased with advancing age.
TTP intensities also generally declined across tissue types when examining
progressively older donors (Table 4, right columns). Representative
micrographs from the GI and immune systems are shown (Figure 5). The dis-agreement
between replicative senescence and in vivo aging was also seen with TTP,
as senescent cells expressed increasingly higher TTP, while advancing age
progressively lowered the number of TTP-expressing cells and TTP abundance per
cell. Although TTP levels can be induced by a variety of stimuli, the
constitutive TTP expression decreased markedly with advancing age.
Table 4. Quantitation of TTP signals in human tissue microarrays.
Shown are the
percentages of positive area (‘% TTP positive') and the signal strength
(‘Intensity') in samples from a range of tissue types and age groups. When
multiple biopsies were quantified in a given tissue and age group, the
average value is shown and the number of tissues examined is indicated in
parenthesis. Values were calculated as explained in the Methods section.
Discussion
Our results reveal an interesting discordance between
the levels of four TTR-RBPs in human fibroblasts undergoing replicative
senescence and their levels in tissues from individuals of increasing age. In
WI-38 cells, senescence potently lowered HuR, AUF1, and TIA-1 levels, while it
increased TTP abundance (Figure 1). Accordingly, we hypothesized that the
levels of HuR, AUF1, and TIA-1 might also decline with aging, while TTP levels
might increase. Using a robust method to quantify immunohistochemical signals
present in different tissue types and age groups, we discovered that in vivo,
these TTR-RBPs were expressed in precisely the opposite pattern: HuR, AUF1, and
TIA-1 remained highly abundant with advancing age, in some cases even
increasing their expression, while TTP levels generally decreased in the aged
groups (compare Figure 1 with Tables 1-4). This discovery was somewhat
surprising, given the wide use of HDFs as an in vitro model of aging and the
broad agreement that senescent cells arise in normal tissues with aging in
vivo, as discussed elsewhere [54]. However, since senescent cells are
terminally arrested and may be cleared by immune cells, perhaps they are
underrepresented in the tissues examined here. Additionally, key differences
exist between cultured HDF senescence and in vivo cellular senescence.
For example, cultured HDFs are exposed to chronic levels of damaging stimuli
such as supraphysiologic oxygen and overabundant growth factors, possibly
triggering a persistent stress response that could elevate TTP levels and lower
HuR, AUF1, and TIA-1 levels. Conversely, a more physiologic setting would
cause stress conditions of different type and magnitude in live organs,
possibly impacting on TTR-RBP abundance. While further experiments are needed
to discern among these possibilities, our findings lead us to join many other
laboratories in questioning the extent to which senescent HDFs recapitulate
features of in vivo aging.
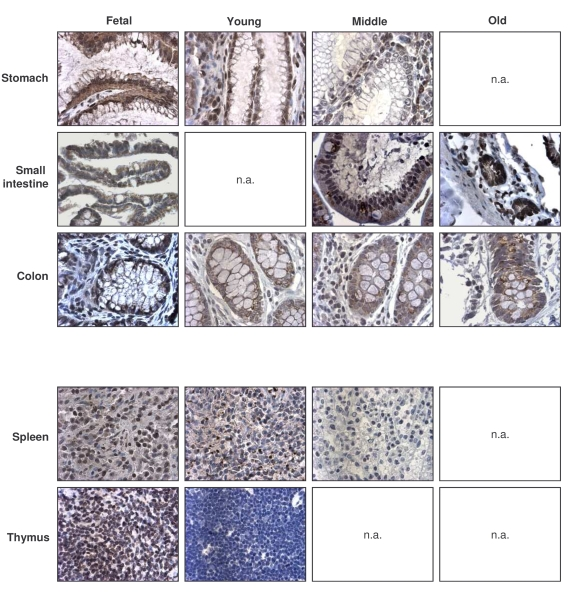
Figure 5. Immunohistochemical detection of TTP across tissue types and age groups. Representative
TTP signals in photomicrographs taken from the indicated tissue sections
from human tissue arrays. Images are shown at ×200 magnification.
A systematic analysis of TTR-RBP expression in human
tissues has not yet been performed. To carry out such an analysis, we obtained
tissue arrays that contained a wide range of human tissue biopsies from
different aged subjects (Methods); in them, we studied TTR-RBP levels using an
immunohistochemical analysis method of color deconvolution that was recently
adapted to tissue array analysis [52,55]. Our
examination of HuR, AUF1, TIA-1, and TTP expression by immunohistochemistry
showed that these proteins were expressed ubiquitously and in high abundance
among many tissues across age groups (Tables 1-4). Lu and Schneider [53] examined systematically the expression of
several TTR-RBPs in adult mice. They reported that HuR was expressed in
numerous tissues, including intestine, thymus, spleen, and liver, while it was
almost undetectable in brain, heart, lung, kidney, and skeletal muscle [53]. This tissue distribution is in agreement with
our findings (Table 1, Figure 2), although in some human tissues, such as liver
and lung, a moderate percentage of cells also expressed HuR, in some cases with
high intensities. The same authors showed that mouse AUF1 was expressed in
highest abundance in thymus and spleen, but was also detectable in brain,
testis, ovary, and uterus, intestine, and lung. Although the levels of AUF1 in
adult brain (M, O) could not be examined, the tissue distribution of AUF1 in
mouse agrees largely with that seen in human. Lu and Schneider used Western
blot analysis to visualize AUF1, which allowed them to examine tissue-specific
differences in isoform abundance [53]. This
assessment was not possible on tissue arrays, since isoform-specific antibodies
for immunohistochemistry are not yet available. However, our analysis yielded
other valuable information, such as the predominantly nuclear localization of
AUF1 and its localization in specific cell types within a given organ (Figures
2-5 and data not shown).
By employing western blot analysis, Beck
and coworkers [56] showed that TIA-1 mRNA and protein were expressed in mouse
brain, spleen, and testis, but not in heart, lung, liver, skeletal muscle, or
kidney. Our results indicate that human TIA-1 was expressed in a more ample range
of tissues, as we also detected high percentages of TIA-1-positive cells in the
GI, urinary, and endocrine systems, and we found generally elevated TIA-1
signals among the different age groups (Table 3). The levels of TTP have also
been examined in adult mouse, with high levels of TTP protein expressed in the
liver, testis, and ovaries, as well as in macrophages [53,57]. In human tissues, we also detect TTP in these organs, but again
find a broader tissue distribution for TTP, with high percentages of cells
expressing TTP and high TTP signals in the urinary and muscular systems, and
especially in the GI tract (Table 4, Figure 5).
HuR has been implicated in numerous cell functions.
Among the four TTR-RBPs studied, HuR is most tightly linked with
proliferation. Binding of HuR to mRNAs encoding cyclin A, cyclin B1, and c-fos
led to their stabilization and/or increased translation, in turn accelerating
cell division [49,58,59]. In keeping with
this function, HuR was low in senescent HDFs (Figure 1) and contributed to
their terminally arrested phenotype [49].
Given this evidence, the finding that HuR was highly expressed in many adult
tissues (M, O) was unexpected. HuR could contribute to the division of epithelial
cells from the GI tract, but it likely does not exert this function in many
other tissues, such as the lung, reproductive organs, and urinary system, which
are populated by many non-dividing cells. Besides proliferation, HuR was shown
to have a broad pro-survival function, by binding to mRNAs encoding
anti-apoptotic proteins like prothymosin α, sirtuin 1 (SIRT1), and bcl-2,
and enhancing their expression [17,60,61]. Additionally, HuR's promotion of
angiogenesis has been linked to its positive influence on the expression of
HIF-1α and VEGF [62,63]. Perhaps the elevated abundance of HuR in
post-mitotic cells helps to carry out an anti-apoptotic function and to ensure
sufficient oxygen supply in terminally differentiated tissues.
All four TTR-RBPs have been linked to the immune
response. HuR function increased following mouse and human activation of
macrophages and T cells [64-67]. In turn, HuR stabilized and/or modulated the
translation of target mRNAs encoding numerous cytokines, such as TNF-α,
IL-6, IL-13, interferon γ, and GM-CSF. AUF1 also targets many of the same
cytokine mRNAs, but it additionally downregulates IL-1β and IL-10 in
immune cells [68-71]. Moreover, as AUF1-knockout mice were unable to degrade
mRNAs encoding proinflammatory cytokines such as TNF-α and IL-1β, LPS
treatment led to severe endotoxic shock [68]. TIA-1 also limits inflammation,
at least in part by binding to the TNF-α mRNA and inhibiting TNF-α
translation. Thus, TNF-α was more highly expressed in macrophages isolated
from TIA-1 knock-out mice than in those isolated from wild type mice [72].
Likewise, TTP limits inflammation by reducing the stability of GM-CSF, IL-2,
and IL-3 mRNAs [44, 73, 74]. Therefore, TTP-/- mice develop severe autoimmune
dysfunction, myeloid hyperplasia, and inflammatory arthritis, due to
deregulated TNF-α and GM-CSF levels [57].
In human immune organs, we observed a strong constitutive presence of HuR, AUF1, and
TIA-1 across the age groups studied, while TTP abundance declined with increasing age. While samples
from the oldest donor group were unavailable on this panel of tissues, our
findings suggest that multiple TTR-RBPs likely contribute to maintaining the
delicate balance that exists between promoting and inhibiting cytokine production.
Taken together, we propose that these TTR-RBPs help to maintain immune
homeostasis throughout human life.
In closing, cancer is among the most prominent
age-related diseases, and there is increasing recognition that TTR-RBPs can
modulate oncogenesis [75, 76]. The pro-malignant influence of HuR and AUF1 is
well established, and numerous cancer-related mRNA targets for these TTR-RBPs
have been identified [15,28]. While TIA-1 can suppress the expression of
cancer-related genes such as COX-2 [36], TIA-1's involvement in cancer is less
well understood. Interestingly, suppression of TTP expression in many cancer
types correlated closely with the tumorigenic phenotype and with patient
prognosis [77], suggesting that TTP could have tumor suppressor function. In
light of our findings that HuR and AUF1 are elevated while TTP levels decline
in tissues from aged donors, we postulate that the higher HuR and AUF1 and
lower TTP could contribute to the increased incidence of cancer seen with
advancing age.
While the links between senescence and aging remain to
be clarified, this analysis reveals interesting distribution patterns for
TTR-RBPs across tissues and age groups. Questions for future consideration
include the influence of tissue type and donor age on the subcellular
localization of TTR-RBPs and their post-translational modification, as these
two parameters profoundly influence the metabolism of target mRNAs. As we work
towards addressing these queries, our findings provide a framework to study the
possible involvement of TTR-RBPs in age-related processes, including the loss
of physiologic function and the onset of diseases associated with advancing
age.
Methods
Cell culture and treatment
. WI-38
human diploid fibroblasts (Coriell Cell Repositories) were maintained in
Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10%
(vol/vol) bovine calf serum (HyClone), 50 μg/ml streptomycin and penicillin,
0.1 mM nonessential amino acids, and 40 μM glutamine in a 5% CO2
incubator.
Western blot analysis
. Whole-cell extracts were prepared as described
previously [61]. Proteins were resolved by 12% sodium dodecyl sulfate
(SDS)-poly-acrylamide gel electrophoresis
and transferred onto polyvinylidene difluoride membranes. Monoclonal antibodies
recognizing HuR (3A2; sc-5261) and GAPDH (6C5; sc-32233) as well as polyclonal
antibodies recognizing TIA-1 (C-20; sc-1751) were from Santa Cruz
Biotechnology; polyclonal antibodies recognizing AUF1 (ab61193) or TTP
(ab33058) were from Abcam. After secondary-antibody incubations, signals were
detected by enhanced chemiluminescence (Amersham).
Immunohistochemistry
. Immunohistochemistry was performed with human adult and fetal normal
tissue (Array II, BioChain Institute, Inc., Hayward, CA, and Pantomics, Inc., San Francisco, CA). The array slides were subjected to heat-induced epitope retrieval,
incubation with primary antibody, and detection with the LSAB+ system (Dako, Carpinteria, CA, USA). A monoclonal anti-HuR antibody (Molecular Probes Inc., Eugene, OR, USA) was used at 0.2 μg/ml. A polyclonal anti-AUF1 antibody (Abcam) was
used at 1:2000 dilution, a polyclonal anti-TIA1 antibody (Santa Cruz) was used
at 1:200 dilution, and a polyclonal anti-TTP antibody (Abcam) was used at
1:1000 dilution.
Slide scanning and image analysis of tissue arrays
. Stained tissue sections were imaged at ×200 total
magnification using a ScanScope CS system (Aperio, Vista, CA). Whole-slide
images were segmented into individual, 24-bit color core image files (TIFF) using
TMALab software (Aperio) for further analysis. Using ImageJ-based macros,
regions of interest (ROI) were selected for each tissue microarray spot to
exclude folded tissues and inappropriate tissue regions [52]. For example, for gastrointestinal tissues, only the
epithelial cell layer was selected as the ROI, while muscular layers were
excluded. Color deconvolution was then used to separate the dye contribution
at each pixel in a given image's ROI; a count of all pixels above an arbitrary
threshold was determined in order to exlude background staining and to
establish a mean threshold of staining. These values were used to generate the
intensity value and to calculate the "% positivity" by dividing the total ROI
pixel count by the DAB positive pixel count in the ROI. The values were further
classified according to age: fetal (F), young (Y, birth to 30
yr-old), middle-aged (M, 30- to 60 yr-old), or old (O, over 60
y), and averaged the scores within each group.
Supplementary Materials
Background immunohistochemical signals in tissue microarrays, without incubating with primary antibody. All other steps were the same as those
used to visualize HuR, AUF1, TIA-1, TTP (Figures 2-5) and prepare Tables 1-4.
Correlation between the percentage positive signals for AUF1 compared with HuR, TIA-1, TTP.
Taking the middle-aged samples, the correlations between
positive signals within a tissue were compared. Correlation
coeficients (R2) indicate that the strongest correlation was seen between HuR and AUF1. In other age groups, AUF1 and HuR also correlated most strongly (not shown).
Collection of tissue biopsies available in both tissue microarrays combined. (M), male; (F), female. y, years old.
Acknowledgments
This research was supported by the National Institute
on Aging-Intramural Research Program, National Institutes of Health. The
authors thank Kristen J. Lecksell (The Johns Hopkins University School of
Medicine) for the digital scanning of the tissue microarray slides.
Conflicts of Interest
The authors of this manuscript have no conflict of
interests to declare.
References
-
1.
Kirkland
JL
, Tchkonia
T
, Pirtskhalava
T
, Han
J
and Karagiannides
I.
Adipogenesis and aging: does aging make fat go MAD.
Exp Gerontol.
2002;
37:
757
-767.
[PubMed]
.
-
2.
Garcia
SN
and Pereira-Smith
O.
MRGing chromatin dynamics and cellular senescence.
Cell Biochem Biophys.
2008;
50:
133
-141.
[PubMed]
.
-
3.
Sedivy
JM
, Banumathy
G
and Adams
PD.
Aging by epigenetics--a consequence of chromatin damage.
Exp Cell Res.
2008;
314:
1909
-1917.
[PubMed]
.
-
4.
Zhang
R
and Zheng
F.
PPAR-γ and aging: one link through klotho.
Kidney Int.
2008;
74:
702
-704.
[PubMed]
.
-
5.
Salih
DA
and Brunet
A.
FoxO transcription factors in the maintenance of cellular homeostasis during aging.
Curr Opin Cell Biol.
2008;
20:
126
-136.
[PubMed]
.
-
6.
Matheu
A
, Maraver
A
and Serrano
M.
The Arf/p53 pathway in cancer and aging.
Cancer Res.
2008;
68:
6031
-6034.
[PubMed]
.
-
7.
Mitchell
P
and Tollervey
D.
mRNA stability in eukaryotes.
Curr Opin Genet Dev.
2000;
10:
193
-198.
[PubMed]
.
-
8.
Orphanides
G
and Reinberg
D.
A unified theory of gene expression.
Cell.
2002;
108:
439
-451.
[PubMed]
.
-
9.
Moore
MJ
From birth to death: the complex lives of eukaryotic mRNAs.
Science.
2005;
309:
1514
-1518.
[PubMed]
.
-
10.
Keene
JD
RNA regulons: coordination of post-transcriptional events.
Nat Rev Genet.
2007;
8:
533
-543.
[PubMed]
.
-
11.
Abdelmohsen
K
, Kuwano
Y
, Kim
HH
and Gorospe
M.
Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence.
Biol Chem.
2008;
389:
243
-255.
[PubMed]
.
-
12.
Hinman
MN
and Lou
H.
Diverse molecular functions of Hu proteins.
Cell Mol Life Sci.
2008;
65:
3168
-31681.
[PubMed]
.
-
13.
Gorospe
M
HuR in the mammalian genotoxic response: post-transcriptional multitasking.
Cell Cycle.
2003;
2:
412
-414.
[PubMed]
.
-
14.
López
de Silanes I
, Zhan
M
, Lal
A
, Yang
X
and Gorospe
M.
Identification of a target RNA motif for RNA-binding protein HuR.
Proc Natl Acad Sci U S A.
2004;
101:
2987
-2992.
[PubMed]
.
-
15.
López
de Silanes I
, Lal
A
and Gorospe
M.
HuR: post-transcriptional paths to malignancy.
RNA Biol.
2005;
2:
11
-13.
[PubMed]
.
-
16.
Kuwano
Y
and Gorospe
M.
Protecting the stress response, guarding the MKP-1 mRNA.
Cell Cycle.
2008;
7:
2640
-2642.
[PubMed]
.
-
17.
Abdelmohsen
K
, Lal
A
, Kim
HH
and Gorospe
M.
Posttranscriptional orchestration of an anti-apoptotic program by HuR.
Cell Cycle.
2007;
6:
1288
-1292.
[PubMed]
.
-
18.
Zhang
W
, Wagner
BJ
, Ehrenman
K
, Schaefer
AW
, DeMaria
CT
, Crater
D
, DeHaven
K
, Long
L
and Brewer
G.
Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1.
Mol Cell Biol.
1993;
13:
7652
-7665.
[PubMed]
.
-
19.
Sarkar
B
, Lu
JY
and Schneider
RJ.
Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family.
J Biol Chem.
2003;
278:
20700
-20707.
[PubMed]
.
-
20.
DeMaria
CT
and Brewer
G.
AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation.
J Biol Chem.
1996;
271:
12179
-12184.
[PubMed]
.
-
21.
Sela-Brown
A
, Silver
J
, Brewer
G
and Naveh-Many
T.
Identification of AUF1 as a parathyroid hormone mRNA 39-untranslated region-binding protein that determines parathyroid hormone mRNA stability.
J Biol Chem.
2000;
275:
7424
-7429.
[PubMed]
.
-
22.
Lal
A
, Mazan-Mamczarz
K
, Kawai
T
, Yang
X
, Martindale
JL
and Gorospe
M.
Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs.
EMBO J.
2004;
23:
3092
-3102.
[PubMed]
.
-
23.
Fialcowitz
EJ
, Brewer
BY
, Keenan
BP
and Wilson
GM.
A hairpin-like structure within an AU-rich mRNA destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics.
J Biol Chem.
2005;
280:
22406
-22417.
[PubMed]
.
-
24.
Xu
N
, Chen
C
and Shyu
A-B.
Versatile role for hnTTR-RBPs in ROS-regulated gene expression 255.
Mol Cell Biol.
2001;
21:
6960
-6971.
[PubMed]
.
-
25.
Raineri
I
, Wegmueller
D
, Gross
B
, Certa
U
and Moroni
C.
Roles of AUF1 isoforms, HuR and BRF1 in AREdependent mRNA turnover studied by RNA interference.
Nucleic Acids Res.
2004;
32:
1279
-1288.
[PubMed]
.
-
26.
Liao
B
, Hu
Y
and Brewer
G.
Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation.
Nat Struct Mol Biol.
2007;
14:
511
-518.
[PubMed]
.
-
27.
Lal
A
, Abdelmohsen
K
, Pullmann
R
, Kawai
T
, Yang
X
, Galban
S
, Brewer
G
and Gorospe
M.
Posttranscriptional derepression of GADD45α by genotoxic stress.
Mol Cell.
2006;
22:
117
-128.
[PubMed]
.
-
28.
Mazan-Mamczarz
K
, Kuwano
Y
, Zhan
M
, White
EJ
, Martindale
JL
, Lal
A
and Gorospe
M.
Identification of a signature motif in target mRNAs of RNA-binding protein AUF1.
Nucleic Acids Res.
2009;
37:
204
-214.
[PubMed]
.
-
29.
Piecyk
M
, Wax
S
, Beck
AR
, Kedersha
N
, Gupta
M
, Maritim
B
, Chen
S
, Gueydan
C
, Kruys
V
and Streuli
M.
TIA-1 is a translational silencer that selectively regulates the expression of TNF-α.
EMBO J.
2000;
19:
4154
-4163.
[PubMed]
.
-
30.
Kandasamy
K
, Joseph
K
, Subramaniam
K
, Raymond
JR
and Tholanikunnel
BG.
Translational control of β2-adrenergic receptor mRNA by T-cell-restricted intracellular antigen-related protein.
J Biol Chem.
2005;
280:
1931
-1943.
[PubMed]
.
-
31.
López
de Silanes I
, Galban
S
, Martindale
JL
, Yang
X
, Mazan-Mamczarz
K
, Indig
FE
, Falco
G
, Zhan
M
and Gorospe
M.
Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1.
Mol Cell Biol.
2005;
25:
9520
-9531.
[PubMed]
.
-
32.
Mazan-Mamczarz
K
, Lal
A
, Martindale
JL
, Kawai
T
and Gorospe
M.
Translational repression by RNA-binding protein TIAR.
Mol Cell Biol.
2006;
26:
2716
-2727.
[PubMed]
.
-
33.
Kim
HS
, Kuwano
Y
, Zhan
M
, Pullmann
RJr
, Mazan-Mamczarz
K
, Li
H
, Kedersha
N
, Anderson
P
, Wilce
MCJ
, Gorospe
M
and Wilce
JA.
Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR.
Mol Cell Biol.
2007;
27:
6806
-6817.
[PubMed]
.
-
34.
Dember
LM
, Kim
ND
, Liu
KQ
and Anderson
P.
Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities.
J Biol Chem.
1996;
271:
2783
-2788.
[PubMed]
.
-
35.
Kawai
T
, Lal
A
, Yang
X
, Galban
S
, Mazan-Mamczarz
K
and Gorospe
M.
Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR.
Mol Cell Biol.
2006;
26:
3295
-3307.
[PubMed]
.
-
36.
Dixon
DA
, Balch
GC
, Kedersha
N
, Anderson
P
, Zimmerman
GA
, Beauchamp
RD
and Prescott
SM.
Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1.
J Exp Med.
2003;
198:
475
-481.
[PubMed]
.
-
37.
Blackshear
PJ
Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover.
Biochem Soc Trans.
2002;
30:
945
-952.
[PubMed]
.
-
38.
Worthington
MT
, Pelo
JW
, Sachedina
MA
, Applegate
JL
, Arse-neau
KO
and Pizarro
TT.
RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein.
J Biol Chem.
2002;
277:
48558
-48564.
[PubMed]
.
-
39.
Brewer
BY
, Ballin
JD
, Fialcowitz-White
EJ
, Blackshear
PJ
and Wilson
GM.
Substrate dependence of conformational changes in the RNA-binding domain of tristetraprolin assessed by fluorescence spectroscopy of tryptophan mutants.
Biochemistry.
2006;
45:
13807
-13817.
[PubMed]
.
-
40.
Carballo
E
, Lai
WS
and Blackshear
PJ.
Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin.
Science.
1998;
281:
1001
-1005.
[PubMed]
.
-
41.
Raghavan
A
, Robison
RL
, McNabb
J
, Miller
CR
, Williams
DA
and Bohjanen
PR.
HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities.
J Biol Chem.
2001;
276:
47958
-47965.
[PubMed]
.
-
42.
Ishmael
FT
, Fang
X
, Galdiero
MR
, Atasoy
U
, Rigby
WF
, Gorospe
M
, Cheadle
C
and Stellato
C.
Role of the RNA-binding protein tristetraprolin in glucocorticoid-mediated gene regulation.
J Immunol.
2008;
180:
8342
-8353.
[PubMed]
.
-
43.
Carrick
DM
and Blackshear
PJ.
Comparative expression of tristetraprolin (TTP) family member transcripts in normal human tissues and cancer cell lines.
Arch Biochem Biophys.
2007;
462:
278
-285.
[PubMed]
.
-
44.
Carballo
E
, Lai
WS
and Blackshear
PJ.
Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability.
Blood.
2000;
95:
1891
-1899.
[PubMed]
.
-
45.
Stoecklin
G
, Ming
XF
, Looser
R
and Moroni
C.
Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway.
Mol Cell Biol.
2000;
20:
3753
-3763.
[PubMed]
.
-
46.
Sawaoka
H
, Dixon
DA
, Oates
JA
and Boutaud
O.
Tristetraprolin binds to the 3'-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site.
J Biol Chem.
2003;
278:
13928
-13935.
[PubMed]
.
-
47.
Stoecklin
G
, Tenenbaum
SA
, Mayo
T
, Chittur
SV
, George
AD
, Baroni
TE
, Blackshear
PJ
and Anderson
P.
Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin.
J Biol Chem.
2008;
283:
11689
-11699.
[PubMed]
.
-
48.
Ogilvie
RL
, Sternjohn
JR
, Rattenbacher
B
, Vlasova
IA
, Williams
DA
, Hau
HH
, Blackshear
PJ
and Bohjanen
PR.
Tristetraprolin mediates interferon-gamma mRNA decay.
J Biol Chem.
2009;
284:
11216
-11223.
[PubMed]
.
-
49.
Wang
W
, Yang
X
, Cristofalo
VJ
, Holbrook
NJ
and Gorospe
M.
Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence.
Mol Cell Biol.
2001;
21:
5889
-5898.
[PubMed]
.
-
50.
Wang
W
, Martindale
JL
, Yang
X
, Chrest
FJ
and Gorospe
M.
Increased stability of the p16 mRNA with replicative senescence.
EMBO Rep.
2005;
6:
158
-164.
[PubMed]
.
-
51.
Lal
A
, Kim
HH
, Abdelmohsen
K
, Kuwano
Y
, Pullmann
R Jr
, Srikantan
S
, Subrahmanyam
R
, Martindale
JL
, Yang
X
, Ahmed
F
, Navarro
F
, Dykxhoorn
D
, Lieberman
J
and Gorospe
M.
p16(INK4a) translation suppressed by miR-24.
PLoS One.
2008;
3:
e1864
[PubMed]
.
-
52.
Cornish
TC
and Halushka
MK.
Color deconvolution for the analysis of tissue microarrays.
Anal Quant Cytol Histol.
2009;
In Press
.
-
53.
Lu
JY
and Schneider
RJ.
Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay.
J Biol Chem.
2004;
279:
12974
-12979.
[PubMed]
.
-
54.
Jeyapalan
JC
and Sedivy
JM.
Cellular senescence and organismal aging.
Mech Ageing Dev.
2008;
129:
467
-474.
[PubMed]
.
-
55.
Ruifrok
AC
and Johnston
DA.
Quantification of histochemical staining by color deconvolution.
Anal Quant Cytol Histol.
2001;
23:
291
-299.
[PubMed]
.
-
56.
Beck
AR
, Medley
QG
, O'Brien
S
, Anderson
P
and Streuli
M.
Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR.
Nucleic Acids Res.
1996;
24:
3829
-3835.
[PubMed]
.
-
57.
Taylor
GA
, Carballo
E
, Lee
DM
, Lai
WS
, Thompson
MJ
, Patel
DD
, Schenkman
DI
, Gilkeson
GS
, Broxmeyer
HE
, Haynes
BF
and Blackshear
PJ.
A pathogenetic role for TNF-α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency.
Immunity.
1996;
4:
445
-454.
[PubMed]
.
-
58.
Wang
W
, Caldwell
MC
, Lin
S
, Furneaux
H
and Gorospe
M.
HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation.
EMBO J.
2000;
19:
2340
-2350.
[PubMed]
.
-
59.
Kim
HH
, Abdelmohsen
K
, Lal
A
, Pullmann
R Jr
, Yang
X
, Galban
S
, Srikantan
S
, Martindale
JL
, Blethrow
J
, Shokat
KM
and Gorospe
M.
Nuclear HuR accumulation through phosphorylation by Cdk1.
Genes Dev.
2008;
22:
1804
-1815.
[PubMed]
.
-
60.
Lal
A
, Kawai
T
, Yang
X
, Mazan-Mamczarz
K
and Gorospe
M.
Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha.
EMBO J.
2005;
24:
1852
-1862.
[PubMed]
.
-
61.
Abdelmohsen
K
, Pullmann
R Jr
, Lal
A
, Kim
HH
, Galban
S
, Yang
X
, Blethrow
JD
, Walker
M
, Shubert
J
, Gillespie
DA
, Furneaux
H
and Gorospe
M.
Phosphorylation of HuR by Chk2 regulates SIRT1 expression.
Mol Cell.
2007;
25:
543
-557.
[PubMed]
.
-
62.
Galbán
S
, Kuwano
Y
, Pullmann
R Jr
, Martindale
JL
, Kim
HH
, Lal
A
, Abdelmohsen
K
, Yang
X
, Dang
Y
, Liu
JO
, Lewis
SM
, Holcik
M
and Gorospe
M.
RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha.
Mol Cell Biol.
2008;
28:
93
-107.
[PubMed]
.
-
63.
Levy
NS
, Chung
S
, Furneaux
H
and Levy
AP.
Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR.
J Biol Chem.
1998;
273:
6417
-6423.
[PubMed]
.
-
64.
Atasoy
UX
and Stellato
C.
Posttranscriptional regulation of IL-13 in T cells: role of the RNA-binding protein HuR.
J Allergy Clin Immunol.
2008;
121:
853
-859.
[PubMed]
.
-
65.
Xu
YZ
, Di Marco
S
, Gallouzi
I
, Rola-Pleszczynski
M
and Radzioch
D.
RNA-binding protein HuR is required for stabilization of SLC11A1 mRNA and SLC11A1 protein expression.
Mol Cell Biol.
2005;
25:
8139
-8149.
[PubMed]
.
-
66.
Casolaro
V
, Fang
X
, Tancowny
B
, Fan
J
, Wu
F
, Srikantan
S
, Asaki
SY
, De
Fanis U
, Huang
SK
, Gorospe
M
, Atasoy
UX
and Stellato
C.
Posttranscriptional regulation of IL-13 in T cells: role of the RNA-binding protein HuR.
J Allergy Clin Immunol.
2008;
121:
853
-859.
[PubMed]
.
-
67.
Johann
AM
, Weigert
A
, Eberhardt
W
, Kuhn
AM
, Barra
V
, von
Knethen A
, Pfeilschifter
JM
and Brüne
B.
Apoptotic cell-derived sphingosine-1-phosphate promotes HuR-dependent cyclo-oxygenase-2 mRNA stabilization and protein expression.
J Immunol.
2008;
180:
1239
-1248.
[PubMed]
.
-
68.
Lu
JY
, Sadri
N
and Schneider
RJ.
Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs.
Genes Dev.
2006;
20:
3174
-3184.
[PubMed]
.
-
69.
Shen
ZJ
, Esnault
S
and Malter
JS.
The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils.
Nat Immunol.
2005;
6:
1280
-1287.
[PubMed]
.
-
70.
Paschoud
S
, Dogar
AM
, Kuntz
C
, Grisoni-Neupert
B
, Richman
L
and Kühn
LC.
Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1.
Mol Cell Biol.
2006;
26:
8228
-8241.
[PubMed]
.
-
71.
Sarkar
S
, Sinsimer
KS
, Foster
RL
, Brewer
G
and Pestka
S.
AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes.
J Interferon Cytokine Res.
2008;
28:
679
-691.
[PubMed]
.
-
72.
Phillips
K
, Kedersha
N
, Shen
L
, Blackshear
PJ
and Anderson
P.
Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis.
Proc Natl Acad Sci U S A.
2004;
101:
2011
-2016.
[PubMed]
.
-
73.
Stoecklin
G
, Lu
M
, Rattenbacher
B
and Moroni
C.
A constitutive decay element promotes tumor necrosis factor alpha mRNA degradation via an AU-rich element-independent pathway.
Mol Cell Biol.
2003;
23:
3506
-3515.
[PubMed]
.
-
74.
Ogilvie
RL
, Abelson
M
, Hau
HH
, Vlasova
I
, Blackshear
PJ
and Bohjanen
PR.
Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay.
J Immunol.
2005;
174:
953
-961.
[PubMed]
.
-
75.
Nguyen-Chi
M
and Morello
D.
Aberrant regulation of mRNA 3' untranslated region in cancers and inflammation.
Med Sci (Paris).
2008;
24:
290
-296.
[PubMed]
.
-
76.
López
de Silanes I
, Quesada
MP
and Esteller
M.
Aberrant regulation of messenger RNA 3'-untranslated region in human cancer.
Cell Oncol.
2007;
29:
1
-17.
.
-
77.
Brennan
SE
, Kuwano
Y
, Alkharouf
N
, Blackshear
PJ
, Gorospe
M
and Wilson
GM.
The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis.
Cancer Res.
2009;
69:
5168
-5176.
[PubMed]
.