The Janus face of DNA methylation in aging
Abstract
Aging is arguably the most familiar yet least-well understood aspect of human biology. The role of epigenetics in aging and age-related diseases has gained interest given recent advances in the understanding of how epigenetic mechanisms mediate the interactions between the environment and the genetic blueprint. While current concepts generally view global deteriorations of epigenetic marks to insidiously impair cellular and molecular functions, an active role for epigenetic changes in aging has so far received little attention. In this regard, we have recently shown that early-life adversity induced specific changes in DNA methylation that were protected from an age-associated erasure and correlated with a phenotype well-known to increase the risk for age-related mental disorders. This finding strengthens the idea that DNA (de-)methylation is controlled by multiple mechanisms that might fulfill different, and partly contrasting, roles in the aging process.
Although age is by far the
biggest risk factor for a wide range of clinical conditions that are prevalent
today, old-age survival has increased substantially during the past half
century. Baby boom generations are growing older, the chance of surviving to
old age is increasing, and the elderly are living longer due to remarkable,
though largely unexplained, reductions in mortality at older ages [1]. Not
surprisingly, these puzzling biodemographic trajectories are difficult to
reconcile with present theories about aging. A key assumption underlying the
theory of evolution holds that fertility and survival schedules are fixed
− a questionable premise for most species in the wild that have evolved
alternate physiological modes for coping with fluctuating environmental
conditions including dauer states (C. elegans), stationary phase
(yeast), diapause (certain insects) and hibernation. Furthermore, studies in
social insects, particularly the honey-bee, have revealed that the same genome
can be alternatively programmed to produce short-lived workers or long-lived
queens. By and by we are coming to realize that the evolution of
whole organisms' traits (birth sizes, growth rates, age and size at maturity,
reproductive investment, mortality rates and lifespan) is crucially shaped by
the interaction of intrinsic and extrinsic factors. How the genetic blueprint
and environmental influences interact with each other is of utmost interest
especially in aging research. Many lines of evidence, including large
epidemiological and extensive clinical and experimental studies, support the
notion that early life events strongly influence later susceptibility to
chronic diseases and mortality rates. An increased understanding of the ability
of an organism to develop in various ways (developmental plasticity), depending
on a particular environment or settings, provides a conceptual basis for these
observations and current biodemographic trends [2, 3].
Developmental plasticity
requires stable modulation of gene expression, and this appears to be mediated,
at least in part, by epigenetic processes such as DNA methylation and histone
modification. This concept entails, however, the question of whether those
epigenetic marks relate to age-associated declines in molecular and cellular
functions. Indeed, the current literature favors the view that epigenetic
mechanisms such as DNA methylation deteriorate with age and may even accelerate
the aging process [4]. A relationship between DNA methylation and aging was
originally proposed in a pioneering study by Berdyshev [5], which showed that
genomic global DNA methylation decreases with age in spawning humpbacked
salmon. In support of this finding, a
gradual global loss of cytosine methylation has been detected in various mouse,
rat and human tissues [6, 7, 8]. Moreover, different types of interspersed
repetitive sequences, which make up a major fraction of mammalian genomes,
appear to be targeted at different ages and to varying degrees by
age-associated hypomethylation [9]. This finding is compatible with the
presence of several mechanisms that regulate global hypomethylation and
possibly contribute at different steps to the aging process.
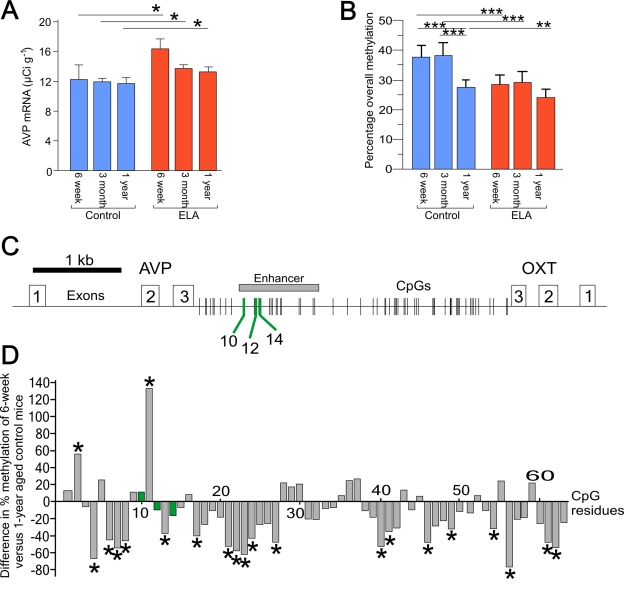
Figure 1. Age related changes in AVP expression and DNA methylation. (A) Aging does not affect AVP mRNA expression in control mice. Early-life
adversity (ELA) leads to a persistent increase in AVP mRNA expression. *P
< 0.05. (B) Age-dependent hypo-methylation occurs only in the
control mice. Early-life adversity leads to a persistent hypomethylation
across the enhancer region in 6-week old mice. **P < 0.005 and ***P <
0.0001. (C) Schematic diagram of the AVP and oxytocin genes
orientated tail-to-tail and separated by the intergenic region (IGR). Exons
are indicated by open numbered boxes and distribution of CpG residues is
shown. The downstream enhancer is boxed in gray with MeCP2 DNA-binding
sites (CpG10, 12, and 14) indicated by green lines. (D) Comparison
of the methylation status of all CpGs in the IGR between 6-week and 1-year
aged control mice shows that the majority of CpGs in the control mice
undergo hypomethylation. In contrast, those methylation landmarks mapping
to MeCP2 DNA-binding sites (marked in green) are protected from
age-associated changes in DNA methylation.
Aside
from global hypomethylation, a number of specific loci have been reported to
become hypermethylated during aging (the ribosomal gene cluster, the estrogen
receptor, insulin growth factor, E-cadherin, c-fos etc.; reviewed in [10]). In
general terms, age-associated hypermethylation is thought to preferentially
affect loci at CpG islands, while loci devoid of CpG islands loose methylation
with age. In addition, a study in humans has revealed that intra-individual
changes in DNA methylation show some degree of familial clustering, indicative
of a genetic component [11].
Taken together, these results
seem to imply that early-life induced programming − in so far that it
relies on DNA methylation − is at a considerable risk to become
insidiously disrupted during aging. This erasing might curtail any long-lasting
programs derived from early-life conditions.
A recent study in mice sheds
new light on this topic. Murgatroyd and coworkers [12] showed that early-life adversity (daily 3-hour separation of
mouse pups from their mother during postnatal days 1−10) caused
persistent hypomethylation at a discrete region of the arginine vasopressin (AVP)
gene enhancer (Figure 1C) in the hypothalamic nucleus paraventricularis (PVN).
This led to a sustained overexpression of AVP (Figure 1A), a key
activator of the hypothalamo-pituitary adrenal (HPA) stress axis. As a result,
early-life adversity evoked a lifelong elevation in glucocorticoid (GC)
secretion, heightened endocrine responsiveness to stressors, reduced stress
coping ability and memory deficits. All of these neuroendocrine and behavioral
alterations are well-known risk factors for aging and frequent features of
age-associated brain pathologies such as major depression and dementia (for
review [13, 14]).
The AVP enhancer is located
downstream of the AVP gene in the intergenic region (IGR) separating the AVP and oxytocin
genes (Figure 1C). Analysis of overall CpG
methylation across the AVP enhancer revealed that the early-life
adversity-induced hypomethylation was strongest at 6 weeks of age though less
prominent in 1-year aged mice compared to controls (Figure 1B). In contrast,
control mice alone showed a clear decrease in methylation at 1 year of age even
though AVP mRNA levels remained unaltered (Figure 1A). This finding suggests
that early-life adversity-induced hypomethylation correlated functionally with
increased AVP transcription and persisted over time, while age-associated
hypomethylation of the AVP enhancer in control mice lacked per se a
functional correlate. This puzzling constellation led the authors to
hypothesize that single CpG residues at the AVP enhancer behaved differentially
with respect to early-life versus age-associated hypomethylation. To elucidate
the cause of such functional heterogeneity among CpG residues at the AVP
enhancer, they went on to correlate CpG methylation across the entire enhancer
with transcriptional activity of the AVP gene. This allowed the
identification of a number of CpG residues (CpG10 and CpGs 12-15 dubbed
'methylation landmarks') that strongly correlated with AVP transcription affinity
DNA-binding sites of the epigenetic reader and writer MeCP2 (methyl-CpG-binding
protein 2)(Figure 2C). MeCP2 serves as a platform upon which synergistic
crosstalk between histone deacetylation, H3K9 methylation and DNA methylation
is played out to confer transcriptional repression and gene silencing (for an
in depth discussion of MeCP2's role in AVP regulation see [15]).
A comparison of the
methylation status of all CpG residues in the IGR in 6-week and 1-year aged
control mice showed that those CpG residues mapping to MeCP2 DNA-binding sites
(marked in green) did not change in the degree of their methylation (Figure 1D). In contrast, 30% of the remaining CpG residues underwent a significant
age-related hypomethylation, while only very few CpG residues (3%) showed a
significant increase. As noted before, this age-associated hypomethylation did
not trigger per se enhanced AVP gene expression.
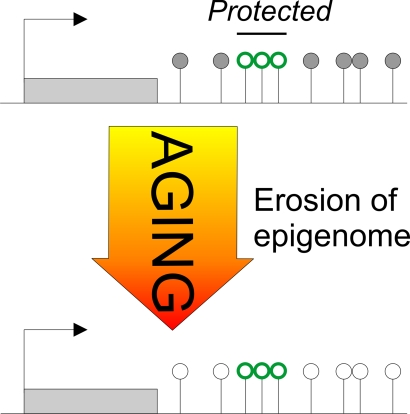
Figure 2. The Janus face of DNA methylation in aging.
Early-life adversity-induced hypomethylation centers on CpG residues mapping to
DNA-binding sites of the epigenetic reader and writer MeCP2 (red
lollipops). Once established, these methylation landmarks are maintained
and do not undergo further age-associated changes in methylation. In
contrast, age-associated hypomethylation maps across the entire AVP locus
without any obvious pattern or preference for potential DNA-binding sites
(black and white lollipops). In this regard, age-associated hypomethylation
appears to behave stochastically, while early-life adversity is targeted.
Taken together, AVP
exemplifies an unexpected double-faced role of DNA methylation in aging.
Hereby, specific environmental stimuli (such as early-life adversity) can induce
site-specific changes in DNA methylation at critical regulatory sites that
underpin sustained changes in gene expression subsequently influencing the risk
of age-associated pathologies. These epigenetic changes are actively controlled
and couple to specific stimuli targeting distinct genes. Due to active
maintenance mechanisms (albeit this does not exclude their extinction by
compensatory or counteracting processes) these epigenetic marks are largely
protected from age-associated changes in DNA methylation (Figure 2).
It
appears that age-associated genome-wide and site-specific (de-)methylation can
indistinguishably disrupt gene expression profiles and lead to the
deterioration of cellular functions. These processes seem to be independent of
a specific stimulus during a critical time window and take place in multiple,
unrelated species. Despite some preliminary evidence from humans that
structural criteria of the DNA (CpG island or the type of repetitive element) age-associated
changes in methylation remain enigmatic. Importantly, however, age-associated
changes in methylation do not inevitably override early life-induced epigenetic
programming (in fact, age-associated hypomethylation of the AVP enhancer
had no effect on mRNA expression levels) and strengthen the idea that these two
processes are functionally and mechanistically distinct. Further research will
be needed to substantiate this concept. However, current work on epigenetic
programming of mice does suggest that differential changes in methylation in
response to early-life adversity and aging apply to other genes in addition to AVP
(Y. Wu, unpublished data). Certainly, the advancement of genome-wide approaches
[16] combining high resolution analysis and functional studies in the field of
epigenetics has the potential to accelerate dramatically our understanding of
the underlying mechanisms in aging and age-associated diseases, ultimately
opening up new possibilities in diagnosis and treatment.
Acknowledgments
This work was supported by
the European Union (CRESCENDO-European Union contract number
LSHM-CT-2005-018652 to D.S.) and the Deutsche Forschungsgemeinschaft (SP 386/4-2 to D.S.).
Conflicts of Interest
The authors in this manuscript have no
conflict of interest to declare.
References
-
1.
Oeppen
J
and Vaupel
JW.
Demography. Broken limits to life expectancy.
Science.
2002;
296:
1029
-1031.
[PubMed]
.
-
2.
Gluckman
PD
and Hanson
MA.
. Living with the past: evolution, development, and patterns of disease.
Science.
2004;
305:
1733
-1736.
[PubMed]
.
-
3.
Gluckman
PD
, Hanson
MA
, Bateson
P
, Beedle
AS
, Law
CM
, Bhutta
ZA
, Anokhin
KV
, Bougnères
P
, Chandak
GR
, Dasgupta
P Smith GD
, Ellison
PT
, Forrester
TE
, Gilbert
SF
, Jablomka
E
, Kaplan
H
, Prentice
AM
, Simpson
SJ
, Uauy
R
and West-Eberhard
MJ.
Towards a new developmental synthesis: adaptive developmental plasticity and human disease.
Lancet.
2009;
373:
1654
-1657.
[PubMed]
.
-
4.
Fraga
MF
and Esteller
M.
Epigenetics and aging: the targets and the marks.
Trends Genet.
2007;
23:
413
-418.
[PubMed]
.
-
5.
Berdyshev
GD
, Korotaev
GK
, Boiarskikh
GV
and Vaniushin
BF.
Nucleotide composition of DNA and RNA from somatic tissues of humpback and its changes during spawning].
Biokhimiia.
1967;
32:
988
-993.
[PubMed]
.
-
6.
Vanyushin
BF
, Nemirovsky
LE
, Klimenko
VV
, Vasiliev
VK
and Belozersky
AN.
The 5¬methylcytosine in DNA of rats. Tissue and age specificity and the changes induced by hydrocortisone and other agents.
Gerontologia.
1973;
19:
138
-152.
[PubMed]
.
-
7.
Wilson
VL
, Smith
RA
, Ma
S
and Cutler
RG.
Genomic 5-methyldeoxycytidine decreases with age.
J Biol Chem.
1987;
262:
9948
-9951.
[PubMed]
.
-
8.
Fuke
C
, Shimabukuro
M
, Petronis
A
, Sugimoto
J
, Oda
T
, Miura
K
, Miyazaki
T
, Ogura
C
, Okazaki
Y
and Jinno
Y.
Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study.
Ann Hum Genet.
2004;
68:
196
-204.
[PubMed]
.
-
9.
Jintaridth
P
and Mutirangura
A.
Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences.
Physiol Genomics.
2010;
[Epub ahead of print]
.
-
10.
Fraga
MF
, Agrelo
R
and Esteller
M.
Cross-talk between aging and cancer: the epigenetic language.
Ann N Y Acad Sci.
2007;
1100:
60
-74.
[PubMed]
.
-
11.
Bjornsson
HT
, Sigurdsson
MI
, Fallin
MD
, Irizarry
RA
, Aspelund
T
, Cui
H
, Yu
W
, Rongione
MA
, Ekstrom
TJ
, Harris
TB
, Launer
LJ; Eiriksdottir G
, Leppert
MF; Sapienza C
, Gudnason
V
and Feinberg
AP.
Intra-individual change over time in DNA methylation with familial clustering.
JAMA.
2008;
299:
2877
-2883.
[PubMed]
.
-
12.
Murgatroyd
C
, Patchev
AV
, Wu
Y
, Micale
V
, Bockmühl
Y
, Fischer
D
, Holsboer
F
, Wotjak
CT
, Almeida
OF
and Spengler
D.
Dynamic DNA methylation programs persistent adverse effects of early-life stress.
Nat Neurosci.
2009;
12:
1559
-1566.
[PubMed]
.
-
13.
De
Kloet ER
, Joels
M
and Holsboer
F.
Stress and the brain: from adaptation to disease.
Nat Rev Neurosci.
2005;
6:
463
-475.
[PubMed]
.
-
14.
Lupien
SJ
, McEwen
BS
, Gunnar
MR
and Heim
C.
Effects of stress throughout the lifespan on the brain, behaviour and cognition.
Nat Rev Neurosci.
2009;
10:
434
-445.
[PubMed]
.
-
15.
Murgatroyd
C
, Wu
Y
, Bockmühl
Y
and Spengler
D.
Genes learn from stress. How infantile trauma programs us for depression.
Epigenetics.
2010;
accepted for publication
.
-
16.
Feinberg
AP
Genome-scale approaches to the epigenetics of common human disease.
Virchows Arch.
2010;
456:
13
-21.
[PubMed]
.