Abstract
N6-methyladenosine (m6A) is one of the most common and well-known internal RNA modifications that occur on mRNAs or ncRNAs. It affects various aspects of RNA metabolism, including splicing, stability, translocation, and translation. An abundance of evidence demonstrates that m6A plays a crucial role in various pathological and biological processes, especially in tumorigenesis and tumor progression. In this article, we introduce the potential functions of m6A regulators, including “writers” that install m6A marks, “erasers” that demethylate m6A, and “readers” that determine the fate of m6A-modified targets. We have conducted a review on the molecular functions of m6A, focusing on both coding and noncoding RNAs. Additionally, we have compiled an overview of the effects noncoding RNAs have on m6A regulators and explored the dual roles of m6A in the development and advancement of cancer. Our review also includes a detailed summary of the most advanced databases for m6A, state-of-the-art experimental and sequencing detection methods, and machine learning-based computational predictors for identifying m6A sites.
Introduction
As is well known, RNAs play an essential role in the translation process, gene regulation, and environmental interactions [1]. Chemical modification is a highly specific and effective method for regulating the functions of biological macromolecules. These macromolecules can be covalently modified after synthesis, such as RNA, DNA, protein, lipids, carbohydrates, and polysaccharides [2]. RNA modification not only has dynamic reversibility but can also be regulated by a wide range of factors, which has led to the emergence of the term “RNA epitranscriptome” and has become increasingly popular in recent years [3]. Recently, more than 100 RNA modification types on bases or ribose have been discovered within all types of RNA, including ribosome RNA (rRNA), transfer RNA (tRNA), messenger RNA (mRNA) and non-coding RNA (ncRNA), which have been confirmed to be involved in many diseases and bio-processes [2, 4, 5]. For example, various RNA modifications in mRNA like N6-methyladenosine (m6A) [6], N4-acetylcysteine (ac4C) [7], N7-methyl guanidine (m7G) [8], 5-methylcytidine (m5C) [9] and N1-methyladenosine (m1A) [10] (Figure 1), are conducive to RNA metabolism steps including splicing, translocation, transcript stability and translation in eukaryotic cells, especially m6A [11]. NAT10 (RNA acetyltransferase)-mediated ac4C can promote mRNA’s stability by extending the mRNA’s half-life in Hela cells [12]. In addition, METTL1-dependent m7G on pre-let-7 regulates pre-let-7 and let-7 expression levels and lung cancer cell migration [13]. Interestingly, even when targeting the same type of methylation, the sets of methyltransferases and demethylases, acting as writers and erasers respectively, can vary across different types of RNA species [14].
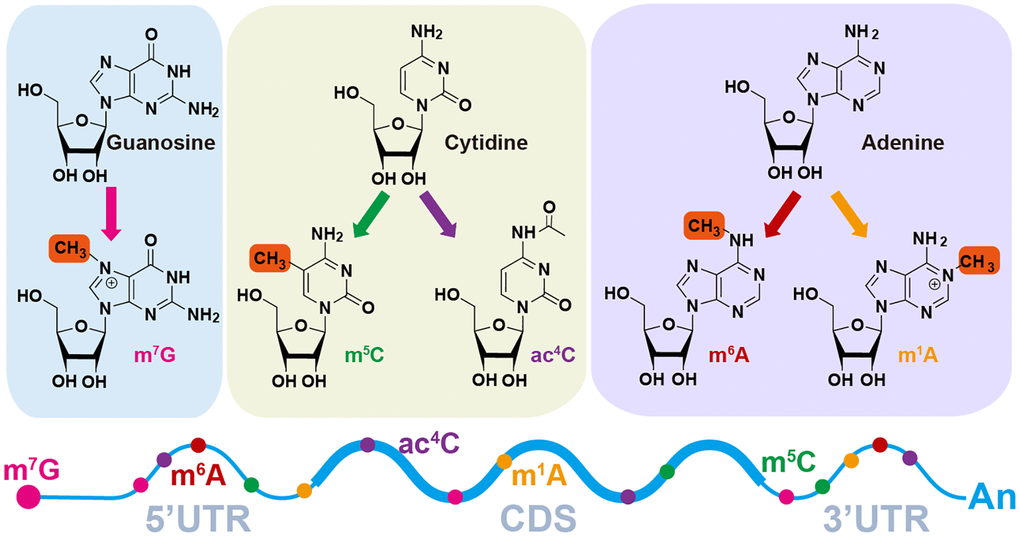
Figure 1. Diverse RNA modification patterns. m7G, N7-methylguanosine; m6A, N6-methyladenosine; ac4C, N4-acetylcytidine; m1A, N1-methyladenosine; m5C, N5-methylcytidine.
This review will summarize the physiological function of m6A or m6A regulators and focus on the interplay of m6A and protein-coding RNA, and ncRNAs (miRNA, circRNA, lncRNA) in various cancers. Finally, we will list the experimental detection and prediction methods of m6A modification residues and summarize m6A-related databases.
Functional mechanism of m6A modification
The enzymes that deposit, remove and bind to RNA modifications are writers, erasers, and readers [3]. Intriguingly, m6A modifications are reversible and dynamic, which is mediated by catalyzing writers and erasers (Table 1). Generally, adenosine methylation occurs preferentially within different consensus sequences. The METTL3/14 complex prefers RRACH (R = G or A; H = A, C or U) in coding regions and 3′UTRs or consensus motif DRACH (D = A, G or U). The crystal structure suggests that another writer, METTL16, prefers a UAC (m6A) GAGAA motif in the bulge of a stem-loop structured RNA (Figure 2, Table 1) [11, 15–17].
Table 1. Biological functions of m6A regulators in RNA metabolism.
| Type | Regulators | Location | Functions | References |
| m6A writers | METTL3 | Nucleus | Installs m6A on mRNAs as well as lncRNAs and mediate RNA translation | [11, 183] |
| METTL14 | Nucleus | Assists METTL3 for m6A deposition and strengthens METTL3 activity | [11, 15, 184] |
| METTL16 | Nucleus | Installs m6A in U6 snRNA | [26] |
| WTAP | Nucleus | Mediates METTL3-METTL14 to the nuclear speckle | [21] |
| VIRMA | Nucleus | Guides the writers to RNA specific region | [23] |
| RBM15 | Nucleus | Binds to m6A complex and recruit it to special RNA site | [22] |
| ZC3H13 | Nucleus | Assists WTAP location to Nito | [185] |
| m6A eraser | FTO | Nucleus | Removes m6A modification and mediates alternative splicing | [27] |
| ALKBH5 | Nucleus | Removes m6A modification | [29] |
| m6A readers | YTHDF1 | Cytoplasm | Promotes RNA translation | [20] |
| YTHDF2 | Cytoplasm | Reduces RNA stability and facilitates RNA degradation | [36] |
| YTHDF3 | Cytoplasm | Mediates the translation with YTHDF1 and strengthens RNA decay with YTHDF2 | [39] |
| YTHDC1 | Nucleus | Promotes RNA alternative splicing and regulate nuclear mRNA transport | [35] |
| YTHDC2 | Cytoplasm | Augments RNA translation | [186] |
| HNRNPA2B1 | Nucleus | Promotes primary microRNA processing and regulates alternative splicing | [42] |
| HNRNPC | Nucleus | Participates in pre-mRNA splicing | [43, 187, 188] |
| HNRNPG | Nucleus | Mediates pre-mRNA splicing | [187] |
| IGF2BP1 | Cytoplasm | Enhances mRNA stability and translation | [47] |
| IGF2BP2 | Cytoplasm | Enhances mRNA stability and translation | [47] |
| IGF2BP3 | Cytoplasm | Enhances mRNA stability and translation | [47] |
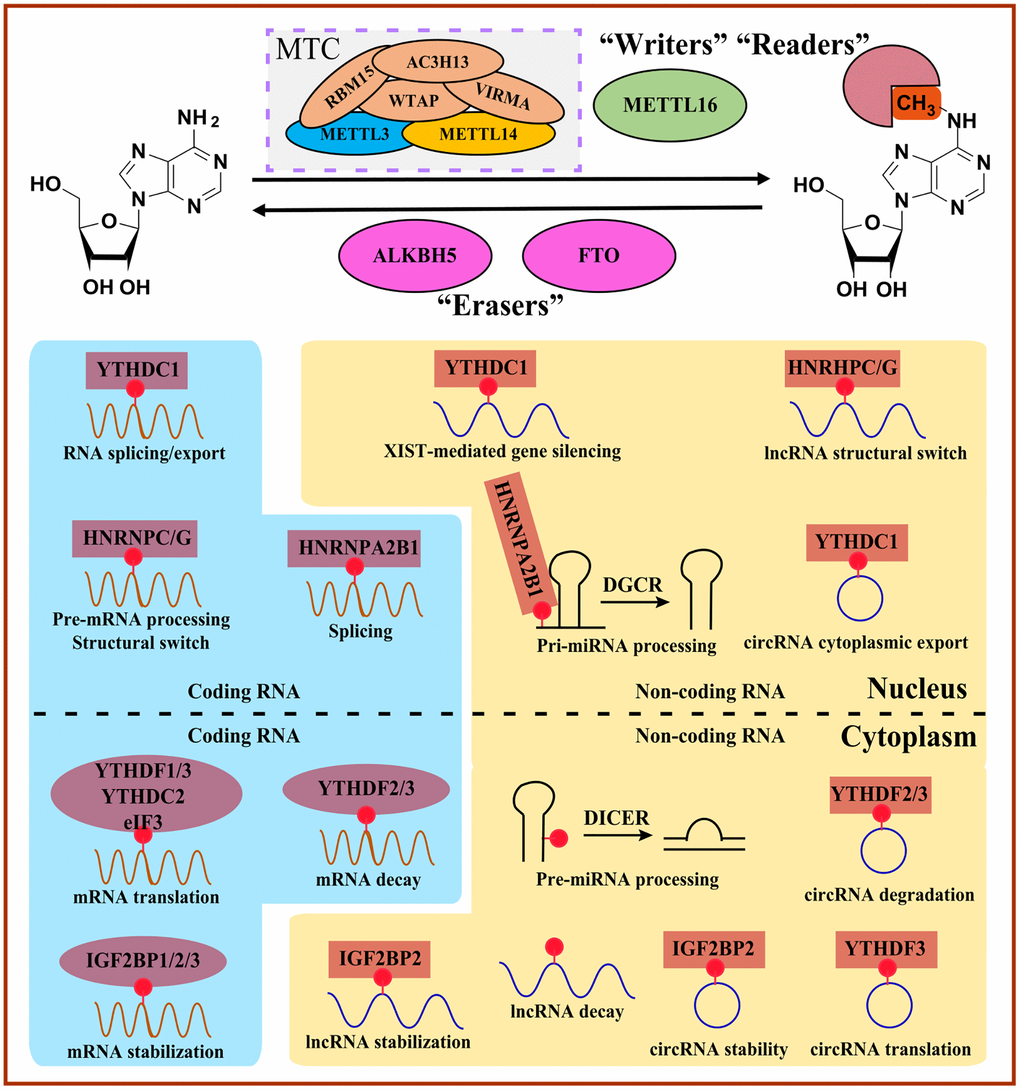
Figure 2. Overview of m6A-mediated RNA metabolism. “Writers” install m6A on coding RNAs and non-coding RNAs, whereas m6A can be reversibly removed by “Erasers”. Diverse m6A “Readers” determine the fate of m6A-modified RNAs involved in the structural switch, splicing, translocation, translation, and decay. MTC, methyltransferase complex.
Writers
The m6A modifications can be catalyzed by m6A writers (m6A methyltransferases). Typically discovered writers include methyltransferase-like 3/14/16 (METTL3/14/16) and other cofactors such as RNA binding motif protein 15/15B (RBM15/15B), wt1-associated protein (WTAP) and vir-like m6A methyltransferase-associated protein (VIRMA), which form the methyltransferase complex (MTC) [11]. Among them, METTL3 is a core component with methyltransferase activity by transferring the methyl from S-adenosyl methionine (SAM) into the N6 site of adenosine [11]. In comparison, METTL14 can impact the stability and methyltransferase activity [18] of the METTL3 complex and recognize the histone modification mark H3K36me3 to regulate the selection of modification sites on RNA [19, 20]. WTAP, known as cofactors and regulatory components of the complex, can guide METTL3/14-RNA binding and locate into nuclear speckles [21]. And RBM15 and its paralog RBM15B show the WTAP-dependent function of recruiting MTC into specific methylate RNA sites [22]. VIRMA is considered an essential part of MTC, showing a similar function as RBM15 [23]. Loss of METTL3 will induce endothelial cells to fail to transform into hematopoietic stem/progenitor cells in zebrafish and mouse embryos [24]. In addition, specific deletion of METTL3 in the central nervous system can lead to severe motif motor dysfunction during lactation and death in mice [25].
Other atypical methyltransferases such as METTL16, ZCCHC4, and METTL5 are discovered and indispensable for m6A [5]. For example, METTL16, a conserved U6 snRNA methyltransferase, regulates the intracellular SAM contents by methylating the SAM synthetase gene MAT2A and participating in the RNA splicing process [26]. Su and colleagues found that METTL16 had the most crucial effect on the survival of tumor cells among all METTL protein families. METTL16 plays a more critical role in almost all tumor cells as an oncogenic gene compared to METTL3 and METTL14. In the nucleus, METTL16 catalyzed the m6A modification of mRNA, especially of nascent mRNA. In the cytoplasm, METTL16 promoted the assembly of 80S ribosomes by directly binding with eIF3a/b and rRNAs, thus improving the translation efficiency of proteins and thus promoting the development of tumors (such as liver cancer) [18]. These results suggest that targeting METTL16, primarily the Mtase domain, may be a potential new tumor therapy strategy.
Erasers
m6A erasers account for the reverse demethylation of modified m6A base, which belongs to the Fe(II)/α-KG-dependent dioxygenase AlkB family: α-ketoglutarate-dependent dioxygenase AlkB homolog 5 (ALKBH5) and the mass-and obesity-associated protein (FTO) [5, 11]. Although FTO is the first identified demethylating enzyme associated with adipogenesis, the following evidence indicates that FTO preferentially demethylates 2’-O-dimethyladenosine (m6Am) [27]. More recently, FTO was revealed to demethylate N1-methyladenosine (m1A) in tRNA [28]. ALKBH5, considered as a secondly discovered m6A demethylase enzyme, has the conservatively catalytic domain to recognize and demethylate m6A modification sites regardless of single or double-stranded RNA [5]. ALKBH5 knockdown in mice affects nascent mRNA synthesis and the splicing rate in mice and humans, respectively [29, 30].
Readers
m6A modification can be recognized by the translation initiation factor eukaryotic initiation factor 3 (eIF3), heterogeneous nuclear ribonucleoproteins (HNRNPs), insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs), and proteins containing the YT521B homology (YTH) domain. In addition, specific m6A readers may mediate different physiological processes, such as immune response and biological rhythm, and participate in RNA metabolism steps such as RNA degradation and translation by binding the m6A modification site directly [11, 31, 32].
Generally, the YTH domain family is composed of YTH domain family protein 1-3 (YTHDF1-3, DF family) and YTH domain-containing protein 1-2 (YTHDC1-2, DC family) in mammalian cells. YTHDC1 belongs to nuclear m6A readers functioning on selectively binding m6A-containing precursor RNAs, while other YTH family members, including YTHDF1, YTHDF2, YTHDF3, and YTHDC2, identified as cytoplasmic m6A readers, regulate the mature mRNAs containing m6A [33].
YTHDC1 also can recognize m6A on XIST and enhance XIST-mediated X chromosome silencing [22]. YTHDC1, in Hela cells, recognizes m6A to form mRNA-protein complex and further interacts with RNA export factor 1 (NXF1) and serine/arginine-rich splicing factor 3 (SRSF3) to facilitate the m6A-methylated mRNAs nuclear export [34]. In addition to the function of silencing and nuclear export, YTHDC1 promotes exon inclusion or exon skipping via selectively recruiting or inhibiting different splicing factors, such as serine-arginine repeat protein. In addition, YTHDC1 recruits competitive RNA splicing factors SRSF3 and blocks serine/arginine-rich splicing factor 10 (SRSF10) from binding to mRNA to regulate exon inclusion and exon skipping, respectively [35].
YTHDC2 preferentially binds m6A within a precisely consensus motif and promotes the translation efficiency of mRNAs. YTHDC2 contains multiple functional elements, such as ankyrin repeats and the R3H domain, and these unique features endow YTHDC2 with many functions, including regulatory effects on RNA binding and structure and recruitment of or binding with other protein complex members [33]. YTHDF1, in Hela cells, can bind to mRNA stop codons and m6A near the 3′UTR region, and augment translation efficiency by interacting with eukaryotic initiation Factor 3 (eIF3) in the 48S translation initiation complex rather than by the m7G-cap-dependent manner [20].
YTHDF2 acts as an m6A reader inducing mRNA degradation [36]. YTHDF2 decodes m6A on more than 2000 mRNAs in HeLa cells. And YTHDF2 knockout can cause the abundance of these mRNAs to increase significantly, suggesting that YTHDF2 is essential for m6A-modified mRNAs [36]. Furthermore, the stability of mRNAs regulated by YTHDF2 positively correlates with the degree of polyadenylation at its 3’end. mRNAs combined with YTHDF2 generally exhibit shorter polyadenylic acid tails than other mRNAs. Typically, YTHDF2 first binds to m6A through the YTH domain, and then its N-terminal domain interacts with the SH (superfamily homolog) domain of CNOT1 and recruits the CCR4/CAF/NOT complex to perform deadenylation [37]. Intriguingly, YTHDF2 is thought to co-localize with both decapping and deadenylation enzyme complexes under normal conditions and directs its target RNAs to processing bodies [37]. However, recent studies showed that almost no significant mRNA degradation events were observed in the p-body, indicating the importance and significance of YTHDF2 in directing the process of RNAs into p-body that still needs to be considered and explored [38]. YTHDF2 can also enhance the expression level of pro-inflammatory cytokines by promoting the degradation of KDM6B, known as the repressive mark H3K27me3 demethylase. Meanwhile, the repressive mark H3K27me3 can inhibit the methylation process on adenosine during the transcription process. KDM6B recruits m6A methyltransferase to remove the H3K27me3 modification near the target gene and promotes the m6A modification on the newly transcribed mRNA, suggesting that complex interactions of m6A modification and histone modification exist and need to be studied [19].
YTHDF3, together with YTHDF1, guides the m6A modified transcript to be combined with YTHDF2 to direct the target RNA into RNA degradation steps. In addition, YTHDF3 could mediate mRNA decay by interacting with YTHDF2. YTHDF3 could cooperate with YTHDF3 to regulate mRNA translation; however, it could not function directly [39]. Moreover, YTHDF3 deficiency will reduce the binding ability of YTHDF1 and YTHDF2 to the target RNA, while YTHDF1 or YTHDF2 loss reduces the amount of RNA that YTHDF3 binds, suggesting that YTHDF3 could indirectly improve m6A-dependent translation efficiency by enhancing the binding ability for YTHDF1 and its target RNAs. Intriguingly, the translation process of circRNA with m6A modification does not depend on YTHDF1 at all, but YTHDF3 is required. Notably, YTHDF3 recruits eIF4G2 that directly binds to internal ribosome entry sites (IRES) to initiate the translation process independent of eIF4E [40, 41].
HNRNP family members, a well-known RNA-binding protein family, involved in m6A modification, consist of heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1), heterogeneous nuclear ribonucleoproteins G (HNRNPG) and heterogeneous nuclear ribonucleoproteins C (HNRNPC). Controversially, is HNRNPA2B1 an m6A reader or not? One study showed that HNRNPA2B1 could directly bind m6A and regulate alternative splicing events by cooperating with METTL3 and primary microRNA processing via interacting with pri-miRNA microprocessor complex component DGCR8. In contrast, a structure-based study revealed an “m6A switch” mechanism rather than specific m6A binding mediated by HNRNPA2B1 [42]. Another two HNRNP family members, HNRNPC and HNRNPG, can regulate the processing of m6A-containing RNA transcripts. HNRNPC also participates in the pre-mRNAs processing steps [43]. However, they do not bind to m6A directly, suggesting that HNRNPC and HNRNPG can bind to the transcripts via RNA structure alternations determined by m6A [44].
eIF3, considered as an m6A reader, recruits 43S protein complex to initiate the translation steps for mRNAs containing 5′UTR m6A without cap-binding factor eIF4E, that is, eIF3 functions in a cap-dependent manner. However, the exact mechanism of eIF3 for m6A recognition is not yet clearly understood. It may rely on the sequence motif like YTH proteins and adjacent RNA structure [45, 46].
IGF2BPs are the conserved m6A-binding proteins, whose RNA-binding domains are composed of RNA recognition motif (RRM) domains and K homology (KH) domains. IGF2BP family members, IGF2BP1, IGF2BP2, and IGF2BP3, can promote the stability and increase the translation efficiency of m6A-modified mRNAs. Additionally, the mRNA stabilization regulated by IGF2BPs may be enhanced by recruiting cofactors such as matrin 3 (MATR3) and ELAV-like RNA binding protein 1 (ELAVL1) [47].
m6A writers, erasers, and readers in cancers
Growing studies revealed that the global abundance of m6A and expression level of m6A regulators often show aberrance across various cancers and are associated with cancer initiation, progression, metastasis, relapse, and therapy [48–50]. Human HCC showed a higher abundance of m6A and augmented gene expression levels when YTHDC2 was knocked down, whereas the global abundance of m6A was reduced significantly in human bladder cancer [51]. This contradiction suggests that research on m6A needs to be more detailed and context dependent. Thus, a comprehensive understanding of m6A regulators may account for better prevention and treatment (Figure 3, Table 2).
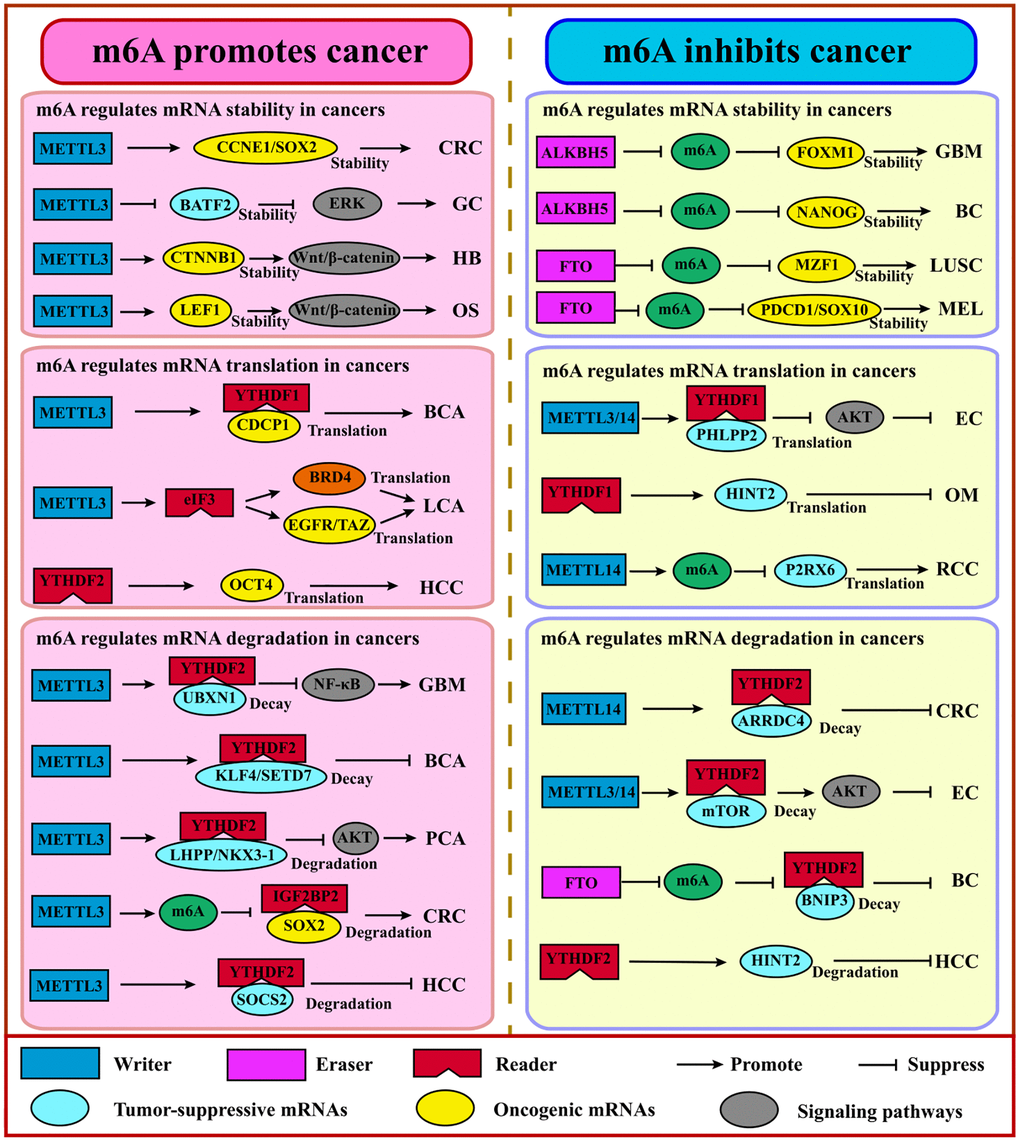
Figure 3. m6A regulates coding RNAs in cancers. Generally, m6A promotes cancer via the stabilization or translation of oncogenes and the degradation of tumor suppressor genes. Adversely, m6A suppresses cancer via the stabilization or translation of tumor suppressor genes, the degradation of the oncogene. Abbreviations: GBM: glioblastoma; GC: gastric cancer; HB: hepatoblastoma; OS: osteosarcoma; BCA: bladder cancer; LCA: lung cancer; HCC: hepatocellular cancer; PCA: prostate cancer; CRC: colorectal cancer; BC: breast cancer; LUSC: lung squamous cell carcinoma; MEL: melanoma; OM: ocular melanoma; RCC: renal cell carcinoma; EC: endometrial carcinoma.
Table 2. Aberrant m6A regulators and its potential mRNA targets in cancers.
| m6A regulator | Cancer type | Regulators Roles | Target | Regulators functions | References |
| METTL3-METTL14 | Acute Myelocytic Leukemia | Oncogene | MYC, SP1, SP2, BCL2, PTEN | ↑Proliferation | [189] |
| Lung cancer | Oncogene | EGFR, BRD4, JUNB | ↑Invasion, ↑Proliferation | [59, 60, 190] |
| Hepatocellular carcinoma | Oncogene | SOCS2, SNAIL | ↑Proliferation, ↑Migration | [54, 191] |
| Endometrial cancer | Tumor suppressor gene | PHLPP2 | ↓Proliferation, ↓Migration | [192] |
| Pancreatic cancer | Tumor suppressor gene | PHLPP2 | ↓Proliferation | [64] |
| Pancreatic cancer | Oncogene | PERP | ↑Growth, ↑Metastasis | [193] |
| Glioblastoma | Oncogene | SOX2, SRSF, ADAR1 | ↑self-renewal, ↑Proliferation | [52, 194, 195] |
| Glioblastoma | Tumor suppressor gene | ADAM19, KLF4, BRCA2, CDKN2A | ↓Growth, ↓self-renewal | [63] |
| Ovarian tumor | Oncogene | AXL | ↑Invasion | [196] |
| Bladder cancer | Oncogene | AFF4, IKBKB, CDCP1, MYC, ALF4 | ↑Growth | [57, 58, 197] |
| Breast cancer | Oncogene | MYC, KLF4, HBXIP | ↑Proliferation, ↑Migration | [198, 199] |
| Breast cancer | Tumor suppressor gene | NOTCH1 | ↓Proliferation | [152] |
| Prostate cancer | Oncogene | LEF1/LHPP | ↑Proliferation, ↑Migration | [200, 201] |
| Colorectal cancer | Tumor suppressor gene | ARRDC4 | ↓Invasion, ↓Migration | [202] |
| Colorectal cancer | Oncogene | CCNE1 | ↑Proliferation | [56] |
| Thyroid carcinoma | Oncogene | TCF1 | ↑Proliferation | [203] |
| Gastric cancer | Oncogene | BATF2, HDGF | ↑Proliferation | [53, 204] |
| Kidney cancer | Tumor suppressor gene | P2RX6 | ↓Invasion, ↓Migration | [205] |
| Hepatoblastoma | Oncogene | CTNNB1 | ↑Proliferation | [55] |
| Nasopharynegal carcinoma | Oncogene | ZNF750 | ↑Growth | [206] |
| WTAP | Hepatocellular carcinoma | Oncogene | ETS1 | ↑Proliferation | [67] |
| Osteosarcoma | Oncogene | HMBOX1 | ↑Tumorigenicity | [207] |
| RBM15 | Laryngeal squamous cell carcinoma | Oncogene | TMBIM6 | ↑Proliferation | [208] |
| FTO | Cutaneous melanoma | Oncogene | PDCD, CXCR4, SOX10 | ↑Tumorigenicity | [74] |
| Acute Myelocytic Leukemia | Oncogene | ASB2, RARA, MYC, CEBPA | ↑Proliferation | [71] |
| Clear cell renal cell carcinoma | Tumor suppressor gene | PGC-1α | ↓Proliferation | [209] |
| Cervical cancer | Oncogene | β-catenin | ↑Development | [210] |
| Lung cancer | Oncogene | MZF1 | ↑Proliferation, ↑Invasion | [211] |
| Hepatocellular carcinoma | Oncogene | PKM2 | ↑Tumorigenicity | [212] |
| Breast cancer | Oncogene | BNIP3 | ↑Growth | [73] |
| ALKBH5 | Glioblastoma | Oncogene | FOXM1 | ↑Tumorigenicity | [80] |
| Acute Myelocytic Leukemia | Oncogene | TACC3, AXL | ↑Self-renewal | [81, 213] |
| Ovarian tumor | Oncogene | NANOG | ↑Proliferation | [214] |
| Pancreatic cancer | Tumor suppressor gene | PER1 | ↓Proliferation | [215] |
| Lung cancer | Oncogene | UBE2C, TIMP3 | ↑Proliferation | [216, 217] |
| Hepatocellular carcinoma | Tumor suppressor gene | LYPD1 | ↓Proliferation | [83] |
| YTHDC2 | Lung adenocarcinoma | Tumor suppressor gene | SLC7A11 | ↓Tumorigenicity | [218] |
| Bladder cancer | Oncogene | CDCP1 | ↑Growth | [197] |
| YTHDF1 | Colorectal cancer | Oncogene | Wnt/β-catenin | ↑Tumorigenicity | [87] |
| Hepatocellular carcinoma | Oncogene | FZD5 | ↑Proliferation | [84] |
| Non-small-cell lung cancer | Oncogene | CDK2, CKD4 | ↑Proliferation | [86] |
| Gastric cancer | Oncogene | FZD7 | ↑Tumorigenicity | [219] |
| Endometrial cancer | Tumor suppressor gene | PHLPP2 | ↓Proliferation | [192] |
| Ovarian tumor | Oncogene | EIF3C | ↑Proliferation, ↑Migration | [220] |
| Ocular melanoma | Tumor suppressor gene | HINT2 | ↓Proliferation | [91] |
| YTHDF2 | Glioblastoma | Oncogene | MYC/UBXN1 | ↑Proliferation, ↑Self-renewal | [221, 222] |
| Ovarian cancer | Oncogene | BMF | ↓Apoptosis | [223] |
| Hepatocellular carcinoma | Oncogene | OCT4 | ↑Growth | [224] |
| Hepatocellular carcinoma | Tumor suppressor gene | EGFR | ↓Proliferation, ↓Growth | [94] |
| Ovarian tumor | Oncogene | FOXK1, PDLIM7 | ↑Growth | [225] |
| Liver cancer/ovarian cancer/lung cancer | Oncogene | E2F1 | ↑Proliferation | [226] |
| IGF2BP1 | Colorectal cancer | Oncogene | SOX2, SEC62 | ↑Tumorigenicity, ↑Proliferation | [227, 228] |
| Hepatocellular carcinoma | Oncogene | SRF/FEN1 | ↑Growth | [225, 226] |
| IGF2BP2 | Clear cell renal cell carcinoma | Oncogene | CDK4 | ↑Proliferation | [229] |
| Colon cancer | Oncogene | CCND1 | ↑Proliferation | [230] |
| IGF2BP3 | Melanoma | Oncogene | MMP2 | ↑Proliferation, ↑Migration | [231] |
| Multiple myeloma | Oncogene | ILF3 | ↑Growth | [232] |
Dysregulation of m6A writers in cancer
METTL3
METTL3 mainly regulates the expression of oncogenes and tumor suppressor genes at the post-transcription level, including mRNA stability and the translation process. METTL3 induces growth enhancement and self-renewal of glioma stem cells. METTL3 is up-regulated in human glioblastoma tissues and induces m6A modification by binding to SOX2 mRNA in 3′UTR. The growth of murine glioblastoma can be decelerated or stagnated by silencing the METTL3 to inhibit the expression level of SOX2 [52]. Additionally, the overexpression of METTL3 and its oncogenic role has been revealed in various cancers such as gastric cancer [53], hepatocellular carcinoma [54], hepatoblastoma [55], colorectal cancer [56], bladder cancer [57, 58] and lung cancer [59, 60]. For example, METTL3 promotes HCC growth and accounts for HCC progression via repressing SOCS2 level. Moreover, higher expression of METTL3 is detected and explored to be associated with CRC metastasis and a poor prognosis [54]. And the higher METTL3 expression in cutaneous squamous cell carcinoma facilitates Np63 expression, which leads to rapid proliferation and growth of CSCC cells [61]. In pancreatic cancer cells, METTL3 knockdown does not affect cell proliferation; however, it increases the sensitivity of cells to anticancer drugs such as gemcitabine and cisplatin [62].
METTL14
Interestingly, METTL14 also plays dual roles in various cancers [52, 63]. The reduction of METTL14 promotes a vicious phenotype by up-regulating oncogene and down-regulating suppressor genes in glioblastoma [63]. Similar results are seen in endometrial cancer: the depletion of METTL14 induces higher cell proliferation by activating AKT pathways [64]. Adversely, METTL14 plays a pro-tumorigenic function in breast cancer by determining the m6A levels of EMT and angiogenesis-related transcripts such as TWIST [65]. In addition, in hematopoietic stem cells and AML, METTL14 deficiency promotes myeloid differentiation and inhibits self-renewal via decreasing m6A signals for targets MYB and MYC to reduce their stability and translation efficiency [66].
WTAP
WTAP is overexpressed in HCC, which enhances tumor cell migration and invasion [67]. Notably, WTAP acts as an oncogene in AML [68]. Another component of the protein complex, VIRMA, can promote cancer cell proliferation and metastasis of breast cancer by regulating CDK1 expression in an m6A-dependent manner [69]. Moreover, in liver cancers, overexpression of VIRMA facilitated cell invasion, and down-regulation of VIRMA inhibited the m6A modification for ID2 or GATA3 mRNAs [70].
Dysregulation of m6A erasers in cancer
FTO
FTO, acting as the m6A demethylase as first discovered, plays an oncogenic role in AML by promoting leukemogenesis via reducing the stability of ASB2 and RARA [71]. R-2HG can inhibit AML progress by targeting and inhibiting FTO to accumulate m6A on MYC and CEBPA transcripts and reduce the stability and expression of their mRNAs [72]. Gathering evidence shows the oncogenic function of FTO in various solid tumors, including breast cancer [73] and melanoma [74]. Additionally, the knockdown of FTO increases m6A methylation on pro-tumorigenic genes, including CXCR4 and SOX10, accounting for increased RNA decay via the YTHDF2 reader [74]. Moreover, reducing FTO can enhance the sensitivity for IFNγ and anti-PD-1 blockade [74]. Xu et al. reported that FTO knockdown activated CD8+ T cells faster and killed tumor cells effectively because FTO could inhibit the activation of CD8+ T cells by regulating m6A signals on the bZIP transcription factors family which mediated the expression of glycolytic genes [75]. MA2, an FTO inhibitor, displayed an intrinsic anti-tumor activity with m6A methylation increasing and cell growth inhibited [76, 77]. However, recent studies showed that FTO acts as a tumor suppressor factor. For instance, Fuks found that down-regulated FTO in epithelial cancers such as lung cancer was associated with a worse prognosis, and FTO deficiency augmented the EMT program through increased m6A signals of key mRNAs within the Wnt signaling cascade [78]. Another study showed that FTO suppressed anchorage-independent growth and reduced apoptosis of ICC cells by impairing oncogene TEAD2 mRNA stability and regulating signaling pathways such as the inflammation pathway and the pyrimidine metabolism pathway [79].
ALKBH5
Unlike FTO expressing widely, ALKBH5 only shows high expression level in testis. ALKBH5 has been shown to promote cell proliferation and metastasis in glioblastoma stem cells [80]. Recent studies reported that ALKBH5 promoted the progress of AML and the self-renewal of leukemia stem cells rather than normal hematopoietic and blood cells [81]. Up-regulated ALKBH5 in epithelial ovarian cancer impairs autophagy and enhances proliferation and invasion [82]. Conversely, ALKBH5 suppresses the malignancy and metastasis of HCC by destabilizing LYPD1 mRNA in an IGF2BP1-dependent manner [83].
Dysregulation of m6A readers in cancer
YTHDF1
More and more evidence supports that YTHDF1 acts as an oncogene by affecting signaling pathways in cancers, including HCC [84], breast cancer [85], NSCLC [86], CRC [87], and Merkel cell carcinoma [88]. For instance, YTHDF1 affects stem cell-like activity and tumorigenicity of CRC cells by mediating the Wnt/β-catenin pathway and regulating the downstream targets FZD9 and WNT6 mRNA [87]. Additionally, oncoprotein c-Myc enhances YTHDF1 expression in colon cancer to induce cancer cell proliferation and resist fluorouracil and oxaliplatin, where YTHDF1 is associated with advanced cancer stages, metastasis, and prognosis [89]. And in NSCLC, the depletion of YTHDF1 will decrease the translation efficiency of m6A-modified transcripts, which inhibits tumor growth under normoxic conditions [86]. And YTHDF1 can recognize and bind to the transcripts marked by m6A that encode lysosomal protease to facilitate antigen degradation, leading to immune escape and incomplete elimination of tumor cells by suppressing the cross-presentation of phagocytic neoantigens and cross-expression of CD8+ T cells [90]. However, in ocular melanoma, YTHDF1, as a tumor suppressor, promotes the translation of HINT2 mRNA (a tumor suppressor) containing m6A [91].
YTHDF2
Growing studies support that YTHDF2 not only plays a pro-tumorigenic role but also serves as a suppressor for various cancers. A higher expression level of YTHDF2 has been found in AML [92] and pancreatic cancer [93], indicating its oncogenic role. Notably, YTHDF2 shortens the half-life of m6A-marked TNF receptor 2 (TNFR2) mRNA, which usually prevents the accumulation of leukemia cells, thus accelerating the spread of AML [92]. Down-regulation of YTHDF2 significantly restrains cell proliferation and migration. Interestingly, YTHDF2 can promote proliferation and inhibit the EMT process and migration by destroying the stability of YAP mRNA, which plays an important role in the Hippo pathway in pancreatic cells [93]. On the other hand, YTHDF2 promotes the degradation of EGFR mRNA by directly binding to the m6A modification site at its 3′UTR to inhibit ERK/MAPK signaling pathway, thereby constraining cell proliferation and growth for HCC cells [94]. Recently, several studies showed that YTHDF2 was associated with an inflammation response. For example, in HCC, the decrease of YTHDF2 can cause inflammation and vascular remodeling, promoting tumor progression. Knocking down YTHDF2 induces a higher abundance of LPS-induced TNF-α, IL-1β, IL-6, and the phosphorylation of NF-κB and MAPK signaling proteins such as p38 and p65 [95].
YTHDF3
So far, the roles and molecular mechanisms of YTHDF3 have yet to be fully elaborated. YTHDF3 can interact with PABP1 and EIF4G2 to promote FOXO3 translation and suppress IFN-stimulated genes which are essential for an anti-viral immune response [96]. In addition, YTHDF3 can recognize m6A-labelled lncRNA GAS5 and mediate its decay to increase YAP expression and promote CRC progression, where GAS5 serves as a driver for YAP nuclear export [97].
YTHDCs
Due to the lesser exploration, YTHDC1 is an alternative splicing regulator. And differential expressed patterns are drawn for YTHDC1. For example, YTHDC1 is highly expressed in colon adenocarcinoma but not in rectal adenocarcinoma, indicating that the region and even the heterogeneity may cause differences in different functions [98]. In regard to YTHDC2, accumulating evidence supports that YTHDC2 induces tumorigenic effects. YTHDC2 expression level is positively correlated with tumor stages in CRC, indicating that YTHDC2 may promote tumor progression [98]. And YTHDC2 strengthens the translation of HIF-1α and TWIST1 by decoding the m6A modification at 5′UTR to promote EMT and cause tumor metastasis [99].
IGF2BPs
IGF2BPs can enhance the stability and translation of mRNA under normal and stress conditions, which can lead to the accumulation of carcinogenic products such as MYC [100]. Knockout of IGF2BP in cervical and liver cancer cells can inhibit cell proliferation and invasion [47]. Additionally, IGF2BP1 overexpression can stabilize PEG10 mRNA with decoding of m6A at 3′UTR to promote endometrial cancer progression [101]. IGF2BP2 can mediate the liver metastasis of colorectal cancer. Mechanistically, YTHDC1 recognizes circNSUN2 m6A modification and forms a circNSUN2/IGF2BP2/HMGA2 ternary complex in the cytoplasm, which promotes the stability of HMGA2 mRNA to function on metastasis [102].
Above all, the m6A regulators, including writers, erasers, and readers, are closely related to the initiation, progression, and metastasis of various cancers by regulating specific downstream RNA targets to determine different RNA processes for oncogenes and tumor suppressor genes, including decay, translation, alternative splicing and nuclear export [11, 17, 103]. And the targeted gene may be involved in apoptosis, autophagy, cell cycle, metabolic programming, angiogenesis, EMT, and others by mediating cancer-associated pathways such as the hippo signaling pathway, NFκB, TGFβ, Hedgehog signaling pathway, Notch signaling pathway etc. [50, 104].
The interplay of m6A and miRNAs in cancers
Usually, miRNAs can regulate gene expression by forming RISC (RNA-induced silencing complex) at the post-transcriptional level with a distributed length of 21–25nt. And miRNAs bind to the 3′UTR of regulated genes, leading to the degradation or translational inhibition of target RNAs. In recent years, growing studies have shown that m6A mediation occurs on miRNAs that affect oncogenes or tumor suppressor genes to impact the tumor process [105, 106] (Figure 4).
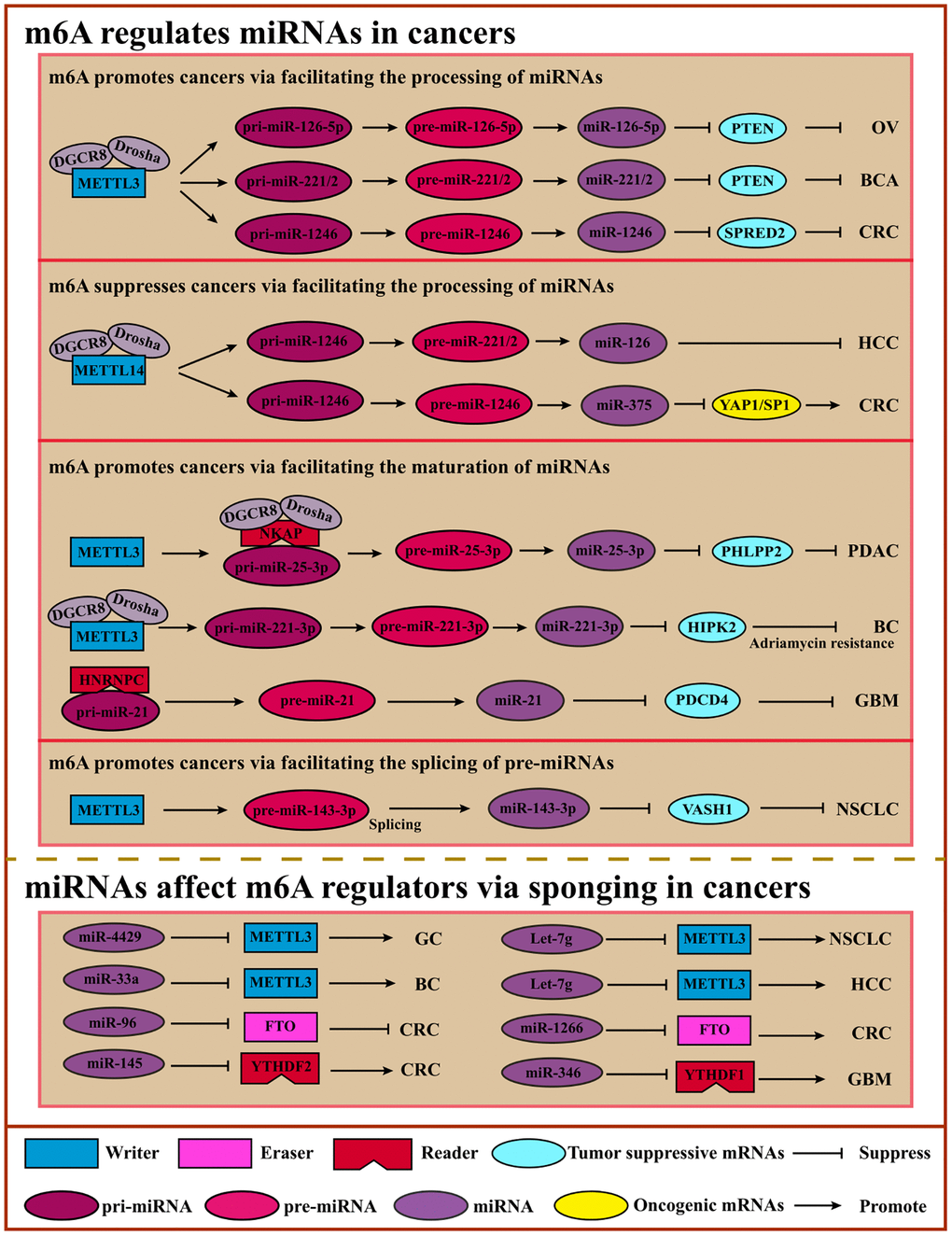
Figure 4. Crosstalk of m6A and miRNAs. m6A can modulate miRNA processing, maturating, or pre-miRNA splicing via “Writers” or “Readers” to inhibit or promote cancers. miRNAs regulate m6A regulators by sponging to affect m6A modification, suppressing or promoting cancers. Abbreviations: OV: ovarian cancer; BCA: bladder cancer; CRC: colorectal cancer; HCC: hepatocellular cancer; PDAC: pancreatic ductal adenocarcinoma; BC: breast cancer; GBM: glioblastoma; NSCLC: non-small cell lung cancer.
M6A methylation affects miRNA processing. For example, miRNA biogenesis involves the following three steps, miRNAs are transcribed as long primary miRNAs (pri-miRNAs) in the nucleus, and these pri-miRNAs are subsequently processed into precursor miRNAs (pre-miRNAs) by a microprocessor complex composed of DGCR8 and DROSHA, and then cleaved by Dicer into mature single-strand miRNAs in the cytoplasm [105].
Intriguingly, the miRNAs biogenesis steps can be affected by m6A regulators in an m6A-dependent manner. METTL3 and HNRNPA2B1 regulate m6A modification on pri-miRNAs to promote the recognition ability of DGCR8 [42, 107]. METTL3 interacts with DGCR8 to promote miR-221/222 maturation which can promote cancer cell proliferation and growth by reducing PTEN expression in bladder cancer cells [108]. In addition, METTL3 also enhances the CRC cells’ metastasis process by facilitating miR-1246 biogenesis and functioning on the miR-1246/SPRED2/MARK signaling pathway [109]. Cigarette smoke condensate promotes the growth of pancreatic ductal adenocarcinoma cells by inducing the maturation of METTL3-mediated m6A-modified pri-miR-25-3p [64]. M6A impacts the maturation of pre-miRNAs and the pre-miRNAs to influence cancer progression. For example, miR-143-3p can be cleaved from pre-miR-143-3p under m6A-modified conditions, playing an essential role in NSCLC with brain metastasis by regulating vasohibin-1 (VASH1) [110]. Like METTL3, METTL14 can also affect miRNA maturation in an m6A-dependent manner to promote or suppress cancer progression. For instance, METTL14 enhances the identification and binding of DGCR8 to facilitate the maturation of pri-miR-126 whose products miR-126 can inhibit HCC cell migration and invasion [111]. Another study showed that METTL14 suppressed CRC progression by promoting the m6A-dependent maturation of miR-375, which functions on YAP1 and SP1 to suppress proliferation and migration [112]. Moreover, m6A reader HNRNPC induces higher expression of miR-21, leading to lower expression of PDCD4 and active migratory and invasive activity in glioblastoma metastasis [113] (Table 3).
Table 3. m6A modulates miRNA processing in cancers.
| m6A regulators | Cancer type | Mechanism | Regulators functions | References |
| METTL3 | Bladder cancer | Promotes the processing of miR-221/222 with DGCR8 | ↑Proliferation | [108] |
| METTL3 | Colorectal cancer | Promotes the processing of miR-1246 with DGCR8 | ↑Migration, ↑Invasion | [109] |
| METTL3 | Pancreatic ductal adenocarcinoma | Promotes miR-25-3p maturation | ↑Migration, ↑Invasion | [64] |
| METTL3 | Non-small cell lung cancer | Promotes splicing of pre-miR-143-3p | ↑Invasion | [110] |
| METTL3 | Ovarian cancer | Promotes miR-126-5p maturation | ↑Migration, ↑Invasion | [233] |
| METTL14 | Hepatocellular carcinoma | Promotes the processing of miR-126 with DGCR8 | ↓Migration, ↓Invasion | [111] |
| METTL14 | Colorectal cancer | Promotes the processing of miR-375 with DGCR8 | ↓Migration, ↓Growth | [112] |
| METTL3 | Breast cancer | Promotes miR-221-3p maturation | ↑Adriamycin resistance | [234] |
| HNRNPC | Glioblastoma | Promotes miR-21 maturation by binding to pri-miR-21 | ↑Migration, ↑Invasion | [64] |
| IGF2BP1 | Hepatocellular carcinoma | Impairs the miRNA-directed decay of the SRF mRNA | ↑Proliferation | [225] |
However, m6A may affect miRNA biogenesis or stability. ALKBH5 interacts with DDX3, which modulates the demethylation of mRNAs by regulating miRNAs [114]. Significantly increased HNONPA2B1 in endocrine-resistant breast cancer reduces the sensitivity to hydro tamoxifen, which is mediated by regulated miRNAs such as miR-29-3p and miR-1268a [115].
Mature miRNAs regulate m6A regulators in cancers. Interestingly, m6A affects the synthesis and function of miRNA directly and impacts the mRNA indirectly mediated by miRNA. IGF2BP2 maintains RAF-1 mRNA stability by obstructing miRNA-mediated degradation, leading to cancer cell proliferation in CRC [116]. Another study showed that miR-1246 suppresses anti-oncogene SPRED2 to induce cancer cell migration and invasion. Mechanistically, METTL3 methylated pri-miR-1246 and facilitated the maturation of pri-miR-1246 [109]. In addition to the above, the m6A regulators may be regulated by miRNA. For example, tumor-suppressive miR-34a suppresses potential oncogenic IGF2BP3 to inhibit cell proliferation and invasion [117]. One other study showed that miR-145 negatively regulated YTHDF2 in HCC cells [118]. However, the mechanism of miRNA functioning in m6A modification remains unclear (Table 4).
Table 4. miRNA regulates m6A regulators via sponging in cancers.
| miRNA | m6A regulators | Cancer type | miRNA functions | References |
| miR-4429 | METTL3 | Gastric cancer | ↓Proliferation, ↑Apoptosis | [235] |
| miR-33a | METTL3 | Non-small cell lung cancer | ↓Proliferation | [236] |
| Let-7g | METTL3 | Breast cancer | ↓Proliferation | [198] |
| miR-600 | METTL3 | Non-small cell lung cancer | ↓Migration, ↑Apoptosis | [237] |
| miR-186 | METTL3 | Hepatocellular carcinoma | ↓Migration, ↓Invasion | [238] |
| miR-96 | FTO | Colorectal cancer | ↑Migration, ↑Invasion | [239] |
| miR-1266 | FTO | Colorectal cancer | ↓Proliferation | [240] |
| miR-744-5p | HNRNPC | Ovarian cancer | ↑Apoptosis | [241] |
| miR-346 | YTHDF1 | Glioma | ↓Proliferation | [242] |
| miR-145 | YTHDF2 | Colorectal cancer/Hepatocellular carcinoma | ↓Proliferation | [118] |
Collectively, m6A is a critical regulator of miRNA at multiple levels, such as maturation, and reverse regulation for m6A regulators by miRNAs is also vital. Therefore, it is essential to investigate the diverse interactions of m6A and miRNAs directly or indirectly.
The interplay of m6A and lncRNAs in cancers
lncRNA, a subgroup of ncRNAs with a length of over 200 nucleotides, can be modified by m6A modification. Researchers are increasingly finding that the interplay of m6A or m6A regulators and lncRNAs could participate in cancer cell proliferation, migration, invasion, or drug resistance. On the one hand, m6A regulators write or recognize the modifiable site lncRNAs which provides to regulate the structure conformation as a structure switch, thereby impacting the RNA-DNA binding ability. On the other hand, lncRNAs serve as endogenous competitive RNAs (ceRNAs) to regulate m6A regulators or directly interact with m6A regulators [119–122] (Figure 5, Tables 5, and 6).
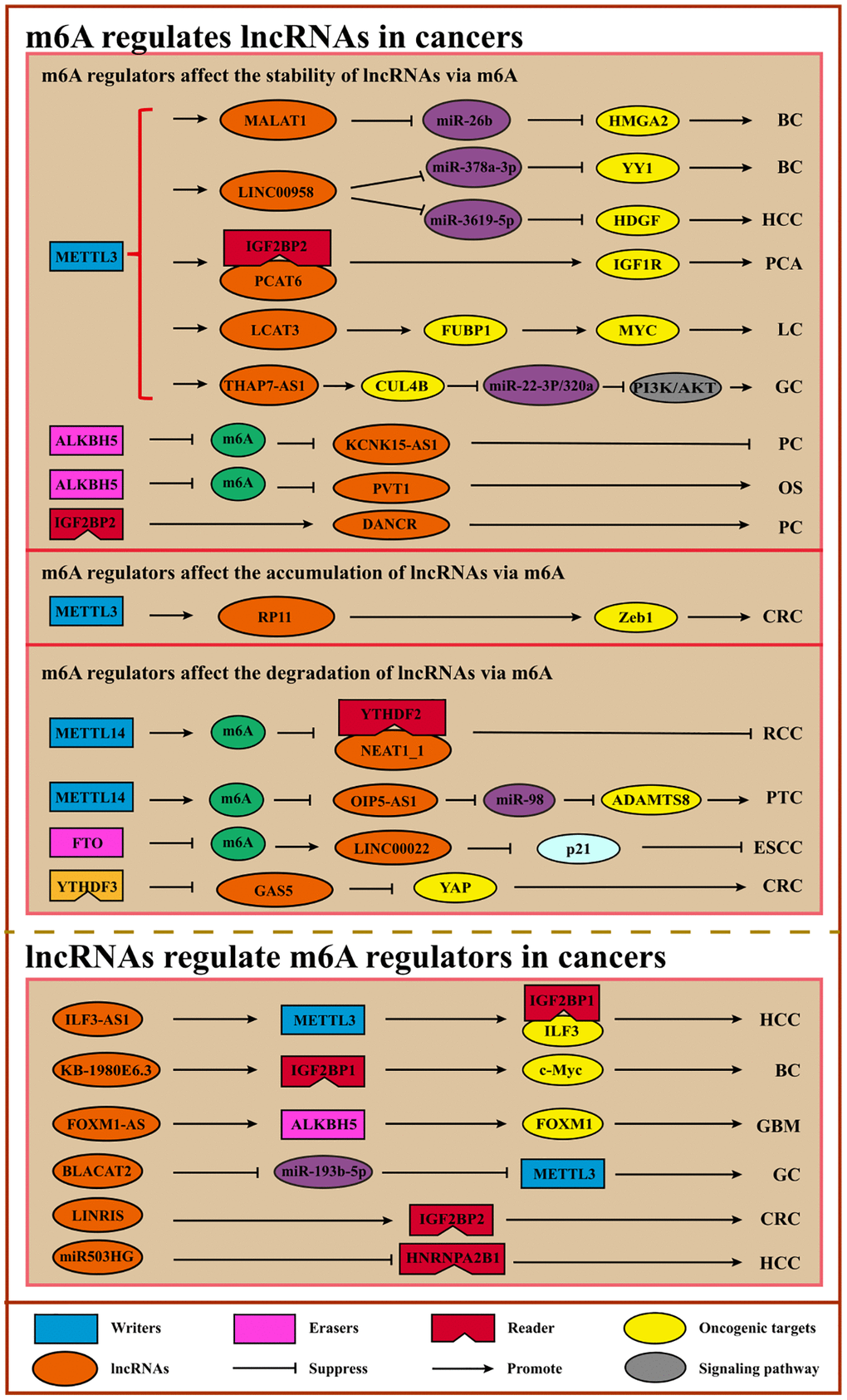
Figure 5. Interplays of m6A and lncRNAs. m6A can modulate the stability, degradation, or accumulation of lncRNAs to regulate cancer initiation, progression, and therapy. Conversely, lncRNAs can serve as ceRNAs to regulate m6A regulators via sponging miRNAs or interact directly with m6A regulators to facilitate or inhibit cancer. Abbreviations: BC: breast cancer; HCC: hepatocellular carcinoma; PCA: prostate cancer; LC: lung cancer; GC: gastric cancer; PC: pancreatic cancer; OS: osteosarcoma; CRC: colorectal cancer; RCC: renal cell carcinoma; PTC: papillary thyroid cancer; ESCC: esophageal squamous cell carcinoma; GBM: glioblastoma.
Table 5. m6A modification regulates lncRNA in cancers.
| m6A regulators | m6A function | LncRNA | Cancer type | Regulator role | LncRNA role | Mechanism | Regulator functions | References |
| METTL3 | writer | MALAT1 | Breast cancer | Oncogene | Oncogene | Promotes RNA stability of MALAT1 | ↑Migration, ↑Invasion | [124] |
| METTL14 | writer | XIST | Colorectal carcinoma | Tumor suppressor gene | Oncogene | Promote degradation of XIST | ↓Migration, ↓Invasion | [123] |
| METTL3 | writer | NEAT1 | Chronic myeloid leukemia | Oncogene | Oncogene | Induces aberrant expression of NEAT1 | ↑Migration, ↑Invasion | [243] |
| ALKBH5 | eraser | PVT1 | Osteosarcoma | Oncogene | Oncogene | Demethylates PVT1 and enhances its expression | ↑Proliferation | [130] |
| METTL3 | writer | LINC00958 | Hepatocellular carcinoma/Breast cancer | Oncogene | Oncogene | Promotes the stability of LINC00958 | ↑Migration, ↑Invasion | [125, 244] |
| METTL3 | writer | FAM225A | Nasopharyngeal carcinogenesis | Oncogene | Oncogene | Promotes the stability of FAM225A | ↑Tumorigenesis, ↑Metastasis | [126] |
| METTL3 | writer | RP11 | Colorectal carcinoma | Oncogene | Oncogene | Increases the accumulation of RP11 | ↑Migration, ↑Invasion | [127] |
| ALHBK5 | eraser | KCNK15-AS1 | Pancreatic cancer | Tumor suppressor gene | Oncogene | Enhances expression of KCNK15-AS1 | ↓Migration, ↓Invasion | [131] |
| FTO | eraser | LINC00022 | Esophageal squamous cell carcinoma | Oncogene | Oncogene | Inhibits the decay of LINC00022 | ↑Proliferation | [132] |
| IGF2BP2 | reader | DANCR | Pancreatic cancer | Oncogene | Oncogene | Promotes the stability of DANCR | ↑Proliferation | [133] |
| METTL3 | writer | ABHD11-AS1 | Non-small cell lung cancer | Oncogene | Oncogene | Promotes the stability of ABHD11-AS1 | ↑Proliferation, ↑Warburg effect | [128] |
| YTHDF3 | reader | GAS5 | Colorectal carcinoma | Oncogene | Tumor suppressor gene | Promotes the degradation of GAS5 | ↑Progression | [97] |
| METTL14 | writer | NEAT1 | Clear cell renal cell carcinoma | Tumor suppressor gene | Oncogene | Promotes degradation of NEAT1_1 | ↓Migration, ↓Invasion | [245] |
| ALKBH5 | eraser | NEAT1 | Colon cancer, gastric cancer | Oncogene | Oncogene | Promotes demethylation of NEAT1 | ↑Migration, ↑Proliferation | [129, 246] |
| METTL3, METTL14 | writer | LNCAROD | Headneck squamous cell carcinoma | Oncogene | Oncogene | Promotes the stability of LNCAROD | ↑Proliferation | [247] |
| METTL3 | writer | FOXD2-AS1 | Cervical cancer | Oncogene | Oncogene | Enhances the stability of FOXD2-AS1 | ↑Tumorigenesis | [248] |
| METTL3 | writer | THAP7-AS1 | Gastric cancer | Oncogene | Oncogene | Enhances the stability of THAP7-AS1 | ↑Migration, ↑Invasion | [249] |
| METTL3 | writer | MEG3 | Hepatocellular carcinoma | Oncogene | Tumor suppressor gene | Promotes degradation of MEG3 | ↑Migration, ↑Invasion | [250] |
| METTL3 | writer | PCAT6 | Prostate cancer | Oncogene | Oncogene | Enhances the stability of PCAT6 | ↑Metastasis | [251] |
| METTL3 | writer | LCAT3 | Lung cancer | Oncogene | Oncogene | Enhances the stability of LCAT3 | ↑Migration, ↑Invasion | [252] |
| METTL14 | writer | Lnc-LSG1 | Clear cell renal cell carcinoma | Tumor suppressor gene | Oncogene | Blocks the interaction of Lnc-LSG1 and ESPR2 | ↓Migration, ↓Invasion | [253] |
| METTL14 | writer | OIP5-AS1 | Papillary thyroid cancer | Oncogene | Tumor suppressor gene | Inhibits OIP5-AS1 expression | ↑Migration, ↑Invasion | [254] |
Table 6. LncRNA modulates m6A regulators in cancer.
| LncRNA | m6A regulators | Type | Cancer type | LncRNA role | Regulator role | Mechanism | References |
| LINC00470 | METTL3 | writer | Gastric cancer | Oncogene | Oncogene | LINC00470 promotes METTL3-mediated PTEN mRNA degradation | [136] |
| LINC00942 | METTL14 | writer | Breast cancer | Oncogene | Oncogene | Recruits METTL14 to mediate CXCR4 and CYP1B1 mRNA stability. | [255] |
| GATA3-AS | KIAA1429 | writer | Hepatocellular carcinoma | Oncogene | Oncogene | Mediate interaction of KIAA1429 with GATA3 pre-mRNA to methylate and degrade GATA3 pre-mRNA | [70] |
| UTM2A-AS1 | METTL3 | writer | Lung adenocarcinoma | Oncogene | Oncogene | Regulates the miR-590-5p/METTL3 axis | [256] |
| LINC00240 | METTL3 | writer | Gastric cancer | Oncogene | Oncogene | Modulates miR-338-5p/METTL3 axis | [257] |
| BLACAT2 | METTL3 | writer | Gastric cancer | Oncogene | Oncogene | Modulates miR-193b-5p/METTL3 | [258] |
| IFL3-AS1 | METTL3 | writer | Hepatocellular carcinoma | Oncogene | Oncogene | Inhibits the degradation of ILF3 mRNA | [259] |
| FOXM1-AS | ALKBH5 | eraser | Glioblastoma | Oncogene | Oncogene | Promotes the interaction of ALKBH5 with FOXM1 to enhance demethylation of FOXM1 | [80] |
| GAS5-AS1 | ALKBH5 | eraser | Cervical cancer | Tumor suppressor gene | Tumor suppressor gene | Interacts with ALKBH5 to demethylate GAS5 to promote tumor suppressor gene GAS5 stability | [260] |
| MALAT1 | IGF2BP2 | reader | Thyroid cancer | Oncogene | Oncogene | miR-204/IGF2BP2/m6A-MYC | [134] |
| LINRIS | IGF2BP2 | reader | Colorectal cancer | Oncogene | Oncogene | LINRIS maintains the stability of IGF2BP2 | [135] |
| KB-1980E6.3 | IGF2BP1 | reader | Breast cancer stem cells | Oncogene | Oncogene | Recruits IGF2BP1 to mediate c-Myc mRNA stability. | [137] |
| LINC00857 | YTHDC1 | reader | Colorectal cancer | Oncogene | Oncogene | Interacts with YTHDC1 to stabilize SLC7A5 | [261] |
| LIN28B-AS1 | IGF2BP1 | reader | Hepatocellular carcinoma, lung adenocarcinoma | Oncogene | Oncogene | Activate LIN28B by interacting with IGF2BP1 | [262] |
| miR503HG | HNRNPA2B1 | reader | Hepatocellular carcinoma | Tumor suppressor gene | Oncogene | Promotes HNRNPA2B1 degradation | [263] |
| DMDRMR | IGF2BP3 | reader | Clear cell renal cell carcinoma | Oncogene | Oncogene | Up-regulates m6A-modified CDK4 via interacting with IGF2BP3. | [264] |
| LINC01234 | HNRNPA2B1 | reader | Non-small-cell lung cancer | Oncogene | Oncogene | Facilitates the processing of pri-miR-106b | [265] |
M6A regulates lncRNA to promote or suppress cancer progression. It has been shown that METTL3, METTL14, and RBM15 can mediate the silencing of lncRNA XIST in a YTHDF2-dependent m6A manner [22, 123]. As for oncogenes, one study showed that METTL3 up-regulated MALAT1 expression to facilitate breast cancer cell migration and invasion via the MALAT1/miR-26b/HMGA2 axis [124]. Moreover, m6A modification on LINC00958 mediated by METTL3 induces its higher expression with its RNA transcripts stabilizing, leading to HDGF up-regulation by sponging miR-3619-59 in HCC [125]. In nasopharyngeal carcinogenesis, METTL3 stabilized FAM225A via m6A modification and up-regulated FAM225A expression, thereby contributing to tumorigenesis and metastasis by sponging miR-590-3p/miR-1275 and up-regulating ITGB3 with the FAK/PI3K/AKT signaling pathway being activated [126]. METTL3 also increases RP11 accumulation via m6A modification in CRC, thus, preventing the degradation of ZEB1 by accelerating the degradation of E3 ligases SIAH1 and FBXO45 [127]. ABHD11-AS1, whose stability can be strengthened by METTL3, promotes NSCLC cell proliferation and the Warburg effect [128]. In addition to the writers, the m6A erasers and readers also regulated lncRNAs. For instance, overexpression of ALHBK5 in GC cells promotes invasion and metastasis by inhibiting NEAT1 methylation [129]. ALHBK5-mediated demethylation of lncRNA PVT1 promotes osteosarcoma cell proliferation [130]. ALHBK5 can also serve as a tumor suppressor. ALHBK5-mediated m6A demethylation on lncRNA KCNK15-AS1 can up-regulate KCNK15-AS1 expression to suppress pancreatic cancer cell migration and invasion while facilitating apoptosis [131]. Another eraser FTO inhibits the LINC00022 decay by decreasing its m6A modification, and LINC00022 promotes ESCC cell proliferation and cell-cycle progression by binding to p21 and promoting its degradation [132]. Regarding readers, IGF2BP2 stabilizes DANCR by recognizing DANCR modification, leading to the proliferation of pancreatic cancer cells [133] (Table 5).
Conversely, lncRNAs can affect m6A via regulating m6A regulators directly or indirectly. For example, MALAT1, considered an oncogene, acts as a ceRNA competitively binding to miR-204 to up-regulate IGF2BP2 expression and enhance MYC expression by m6A modification in thyroid cancer [134]. Moreover, LINRIS maintains the stability of IGF2BP2 by blocking ubiquitination, leading to CRC cell proliferation via MYC-mediated glycolysis [135]. Another study showed that LINC00470 interacted with METTL3 to facilitate PTEN mRNA degradation, thereby accelerating distal migration [136]. LncRNA KB-1980E6.3 is overexpressed and promotes breast cancer stem cell tumorigenesis and self-renewal via recruiting IGFBP1 and retaining c-Myc stability [137]. Furthermore, YTHDF3 accelerates the degradation of GAS5 to regulate YAP signaling and promote CRC progression [97]. Additionally, lncRNA ARHGAP5-AS1 can recruit METTL3 to stimulate ARHGAP5 mRNA m6A modification and stabilize ARHGAP5 mRNA, leading to chemoresistance in gastric cancer [138] (Table 6).
Overall, the interactions between m6A modification or m6A regulators and lncRNAs provide insight for a better understanding of cancer initiation, progression, and therapy.
The interplay of m6A and circRNAs in cancers
Circular RNAs, a group of ncRNAs characterized by covalently closed loop structures with neither 5′ to 3′ polarity nor polyadenylated tail, are more stable than linear RNAs due to their ring-closed structure [139]. Accumulating evidence shows the crosstalk between m6A modification or m6A regulators and circRNAs. Indeed, M6A modification modulates diverse functions of circRNAs, including expression, translation, location, and interaction with RNA or protein. Conversely, circRNAs can also affect m6A modification by regulating the m6A regulators, including writers, erasers, or readers. Surprisingly, aberrant circRNAs have been found and modified by m6A in diverse cancers. Exploring the crosstalk of m6A and circRNA is necessary for cancer therapy [119, 140] (Figure 6, Table 7).
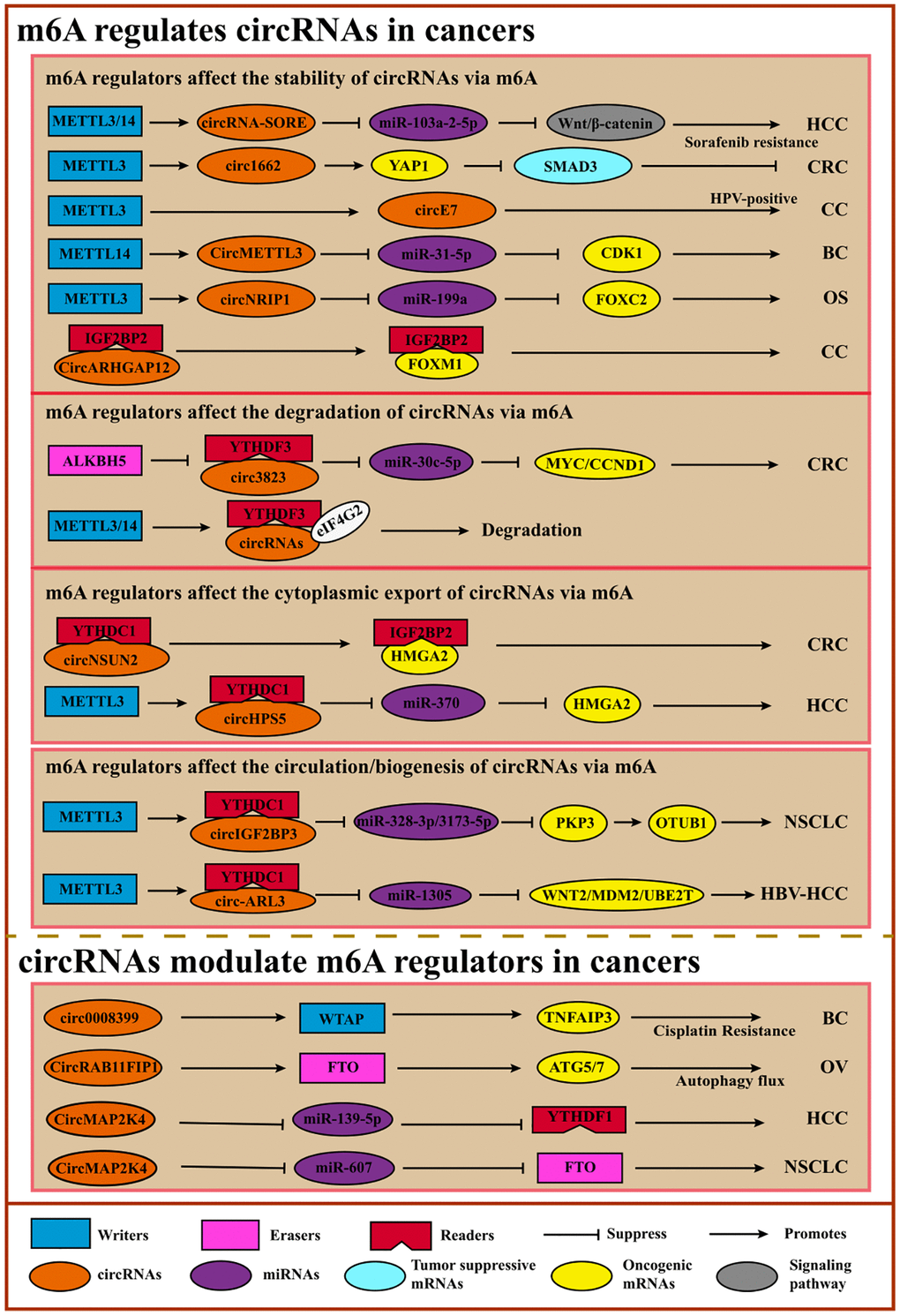
Figure 6. Crosstalk between m6A and circRNAs. m6A facilitates the circulation, accumulation, translocation, translation, or degradation of circRNA to promote or suppress cancers via diverse cancer-associated signaling pathways or tumor-related oncogene/suppressors. On the contrary, circRNAs regulate m6A regulators like lncRNAs.
Table 7. The functions of m6A-modified circRNAs in cancers.
| m6A regulators | m6A function | circRNA | Cancer type | Regulator role | circRNA role | Mechanism mediated by m6A | circRNA Functions | References |
| METTL3 | writer | circNRIP1 | Osteosarcoma | oncogene | oncogene | Promotes circNRIP1 expression | ↑Proliferation, ↑Migration | [141] |
| ALKBH5/YTHDF3 | eraser/reader | circ3823 | Colorectal cancer | tumor suppressor gene | oncogene | Reduces circ3823 stability | ↑Proliferation, ↑Metastasis, ↑Angiogenesis | [142] |
| METTL14/FTO | writer/eraser | circMETTL3 | Breast cancer | oncogene/tumor suppressor gene | oncogene | Promotes/Reduces circMETTL3 expression | ↑Proliferation, ↑Migration, ↑Invasion | [143] |
| YTHDC1 | reader | circNSUN2 | Colorectal cancer | oncogene | oncogene | Facilitates circNSUN2 cytoplasmic export and mediates HMGA2 stabilization | ↑Metastasis | [102] |
| METTL3/YTHDC1 | writer/reader | circHPS5 | Hepatocellular carcinoma | oncogene | oncogene | Facilitates m6A-modified circHPS5 accumulation and cytoplasmic export | ↑Proliferation, ↑Migration | [148] |
| METTL3/14 | writer | circRNA-SORE | Hepatocellular carcinoma | | sustains sorafenib resistance | Increases circRNA-SORE stability | Sustains sorafenib resistance | [266] |
| METTL3/YTHDC1 | writer/reader | circ-ARL3 | HBV- Hepatocellular carcinoma | oncogene | oncogene | Favors reverse splicing and biogenesis of circ-ARL3 | ↑Tumorigenesis | [149] |
| YTHDF2 | reader | circASK1 | lung adenocarcinoma | | ameliorates gefitinib resistance | Increases endoribonucleolytic cleavage of m6A-modified circASK1 | Ameliorate gefitinib resistance, ↑Apoptosis | [267] |
| METTL3/YTHDC1 | writer/reader | circIGF2BP3 | Non-small-cell lung cancer | oncogene | oncogene | Promotes circIGF2BP3 circularization | Facilitate tumor immune evasion | [150] |
| IGF2BP2 | reader | circARHGAP12 | cervical cancer | | oncogene | Promotes circARHGAP12 stability | ↑Progression | [144] |
| | circPVRL3 | Gastric cancer | | tumor suppressor gene | Promotes the circPVRL3 translation | ↓Proliferation, ↓Migration | [151] |
| METTL3 | writer | CircE7 | cervical carcinoma | | oncogene | Promotes CircE7 stability | ↑Growth | [145] |
| METTL3 | writer | circ1662 | Colorectal cancer | oncogene | oncogene | Induces circ1662 expression and install m6A on it | ↑Migration, ↑Invasion | [146] |
| METTL3 | writer | circCUX1 | Hypopharyngeal squamous cell carcinoma | | | Promotes circCUX1 stability and expression | Confers radio-resistance | [268] |
m6A affects the location, biogenesis, stabilization, or translation of circRNAs, leading to cancer proliferation, migration, or invasion [140]. For example, in osteosarcoma, METTL3-mediated m6A modification on circNRIP1 can promote its expression to enhance cell proliferation and migration [141]. M6A-modified Circ3823 contributes to CRC cell proliferation and metastasis, which is regulated negatively by ALKBH5 and YTHDF3 [142]. Another study showed a similar result: high expression of circMETTL3 induced by METTL14-mediated m6A modification leads to breast cancer cell migration and invasion [143]. Other studies also reported that m6A could stabilize the circ-RNAs to impact cancer progressions, such as circARHGAP12 and CircE7 in cervical cancer [144, 145] and circ1662 in CRC [146]. As mentioned above, some studies showed that m6A regulated the translocation of circRNAs. In colorectal cancer, m6A mediation facilitates circNSUN2 cytoplasmic export and further mediates HMGA stabilization to promote cancer metastasis [147]. Another study reported that METTL3 could direct circHPS5 formation and METTL3-controlled m6A could also facilitate the circHPS5 accumulation and cytoplasmic export, indicating that m6A regulators may participate in the formation of circRNAs [148]. Oncogenic circ-ARL3, whose biogenesis is controlled by m6A modification, promotes tumorigenesis in HBV-HCC [149]. Furthermore, METTL3-mediated m6A modification promotes circIGF2BP3 circularization, facilitating NSCLC cells’ immune evasion. Although circRNAs are considered ncRNAs, some circRNAs have the potential to encode short peptides or proteins [150]. M6A promotes circPVRL3 translation, and circPVRL3 can inhibit GC cell migration and invasion [151]. Despite these initial investigations, the mechanisms underlying the functions of m6A-modified circRNAs still need to be explored, especially the translation process (Table 7).
circRNAs can regulate m6A modification on specific target genes by affecting the m6A regulator’s expression or functions. For instance, circNOTCH1 can competitively bind to METTL14 to regulate NOTCH1 mRNA modification, thereby controlling NOTCH1 expression [152]. CircRNAs can also interact with writers or erasers to control target gene m6A modification to further regulate the expression. CircARHGAP12 can interact with IGF2BP2 to accelerate the stability of FOXM1 mRNA, leading to cervical cancer progression [144]. Another study revealed that CircRAB11FIP1 promoted epithelial ovarian cancer cell proliferation and invasion via interacting with FTO to decrease the expression of ATG5 and ATG7 by demethylating their m6A modification [153]. And other circRNAs like circARHGAP12 in cervical cancer [144] and circ0008399 in bladder cancer [154] also perform similar functions. Like lncRNA, circRNAs can also act as ceRNAs to regulate the m6A regulators. For example, CircMAP2K4 and hsa_circ_0072309 enhance HCC and NSCLC cells proliferation and migration via the CircMAP2K4/miR-139-5p/YTHDF1 axis [155] and hsa_circ_0072309/miR-607/FTO axis [156], respectively. Importantly, circRNAs can only regulate m6A modification indirectly via m6A regulators rather than themselves directly (Table 8).
Table 8. CircRNA modulates m6A regulators in cancer.
| circRNA | m6A regulators | m6A function | Cancer type | circRNA role | Mechanism | circRNA Functions | References |
| circARHGAP12 | IGF2BP2 | reader | cervical cancer | oncogene | Interacts with IGF2BP2 to accelerates the stability of FOXM1 mRNA | ↑Progression | [144] |
| CircRAB11FIP1 | FTO | eraser | epithelial ovarian cancer | oncogene | Interacts with FTO to decrease expression of ATG5 and ATG7 | ↑Proliferation, ↑Invasion | [153] |
| circ_KIAA1429 | YTHDF3 | reader | Hepatocellular carcinoma | oncogene | YTHDF3 enhanced Zeb1 mRNA stability | ↑Proliferation, ↑Migration, ↑Invasion | [269] |
| CircMAP2K4 | YTHDF1 | reader | Hepatocellular carcinoma | oncogene | CircMAP2K4/miR-139-5p/YTHDF1 axis | ↑Proliferation | [155] |
| circ0008399 | WTAP | writer | bladder cancer | oncogene | Interacts with WTAP and increase TNFAIP3 stability | Reduce chemosensitivity to CDDP | [154] |
| circNOTCH1 | METTL14 | writer | Non-small-cell lung cancer | oncogene | Regulates NOTCH1 mRNA m6A modification by competing for METTL14 binding | ↑Growth | [152] |
| hsa_circ_0072309 | FTO | eraser | Non-small-cell lung cancer | oncogene | hsa_circ_0072309/miR-607/FTO axis | ↑Tumorigenesis, ↑Invasion | [156] |
| CircMEG3 | METTL3 | writer | liver cancer | tumor suppressor gene | Inhibits the expression of METTL3 dependent on HULC | ↓Growth | [270] |
| circNDUFB2 | IGF2BPs | reader | Non-small-cell lung cancer | tumor suppressor gene | Destabilizes IGFBPs | ↓Growth | [271] |
| CircPTPRA | IGF2BP1 | reader | Bladder cancer | tumor suppressor gene | Blocks the recognition of m6A via interacting with IGF2BP1 | ↓Proliferation, ↓Migration, ↓Invasion | [272] |
m6A databases and m6A sites predictors
To explore the function of m6A systematically and conveniently, the investigator collected and built diverse databases such as m6A-Atlas [157], m6A2Target [158], RMVar [159], and RMDisease [160]. Among them, some summarize the existing m6A modification sites and the potential targets of m6A regulators, such as m6A-Atlas, MET-DB v2.0 [161], RMBase v2.0 [162], and m6A2Target [158]. While some research teams provide the associations between m6A modification sites or m6A-associated variants, m6A-associated histone modification, and chromatin accessibility regions and diseases such as REPIC [163] and RMDisease [160]. And RMDisease summarizes disease-specific genetic variants that affect RNA modification sites (Table 9).
Table 9. m6A-related databases.
| Database | Year | Characteristics | Species | URL | Reference |
| RMBase v2.0 | 2018 | It is a update version with modules “Motif”, “modRBP” and “modMetagene” for visualization of RNA modification sites and explore their potential functions such as SNP or RBP regions. | Human/Mouse/Yeast/Arabidopsis/Zebrafish/Rat/Macaque/Chimpanzee/E.coli/Fruit fly/P.aeruginosa/ Pig/Rhesus | http://rna.sysu.edu.cn/rmbase/ | [162] |
| M6A2Target | 2020 | It provides the potential targets or validated targets for m6A writers, erasers and readers. | Human/Mouse | http://m6a2target.canceromics.org/ | [158] |
| m6A-Atlas | 2020 | It provides a high-confidence collection of m6A modification sites and the quantitative epitranscriptome profiles | Human/Mouse/Rat/Zebrafish/Fly/Arabidopsis/Yeast | http://www.xjtlu.edu.cn/biologicalsciences/atlas | [157] |
| RMVar | 2021 | It provides RNA modification (RM)-associated variants for 9 kinds of RNA modifications and their potential function on posttranscriptional regulation and disease. | Human/Mouse | https://rmvar.renlab.org/ | [159] |
| m6AVar | 2018 | It provides m6A modification sites and potential function of m6A associated variants on post-transcriptional regulation and disease. | Human/Mouse | http://m6avar.renlab.org/ | [273] |
| REPIC | 2020 | It provides m6A modification sites, m6A-associated histone modification and chromatin accessibility regions | Human/Mouse/Yeast/Arabidopsis/Zebrafish/Rat/Macaque/Chimpanzee/E.coli/Fruit fly/P.aeruginosa | https://repicmod.uchicago.edu/repic | [163] |
| MET-DB v2.0 | 2018 | It provides context specific m6A peaks as well as m6A sites at single-base level and the targets of m6A regulators. | Human/Mouse/Rat/Zebrafish/Fly | http://compgenomics.utsa.edu/MeTDB; http://www.xjtlu.edu.cn/metdb2 | [161] |
| CVm6A | 2019 | It provides a visualization interface for m6A patterns in cell lines and their potential function in disease and development. | Human/Mouse | http://gb.whu.edu.cn:8080/CVm6A | [274] |
| m6Acomet | 2019 | It provides gene ontology functions associated m6A sites. | Human | http://www.xjtlu.edu.cn/biologicalsciences/m6acomet | [275] |
| M6ADD | 2021 | It provides analysis interface for m6A modification and the associations between m6A modification and gene disorders and diseases | Human/Mouse | http://m6add.edbc.org/ | [276] |
| RMDisease | 2021 | It provides disease specific genetic variants that affect RNA modifications sites. | Human | http://www.xjtlu.edu.cn/biologicalsciences/rmd | [160] |
| RNAmod | 2019 | It is a web platform for mRNA modifications analysis, annotation and visualization, including m6A distribution, modification coverage and functional ontology. | Human/Mouse/Yeast/Arabidopsis/Zebrafish/Rat/Macaque/Chimpanzee/Fruit fly/Pig | https://bioinformatics.sc.cn/RNAmod | [277] |
| DRUM | 2019 | It is a multi-layer heterogeneous network-based approach by utilizing RWR algorithm to integrate the associations among genes, m6A sites and diseases. | Human/Mouse | http://www.xjtlu.edu.cn/biologicalsciences/drum | [278] |
| RNAWRE | 2020 | It deposits RNA modification effectors | / | http://rnawre.bio2db.com/ | [279] |
Additionally, many tools and platforms predict m6A modification sites based on m6A-seq or MeRIP-seq [164] datasets across diverse species. Generally, the predictors for m6A sites go through the following steps:
Transforming the RNA sequences with/without m6A sites into feature representations, including PseDNC, PseTNC, NCP, ANF, DNC, PSNP, PSDP, NC, PCP, AC, CC, NPPS, STTNC, PSNSP, PSDSP, KSNPF, PS(k-mer) NP, PCPs and RFHC.
Selecting essential features using IFS, RFE, or SFS (optional step).
Training the classifier based on the selected features to generate the model, such as random forest model, SVM, KNN, or deep neural network.
Voting with ensemble classifier (optional step).
Evaluating the performance of the classifiers using a general evaluation index such as ROC.
Notably, it is necessary to establish an evaluation system to evaluate the merits of these software for better supervision and efficiently convenient for users because of too many prediction tools (Table 10).
Table 10. Computational methods for predicting m6A modification from RNA sequences.
| Predictor | Year | Characteristics | Species | URL | Reference |
| SRAMP | 2016 | It integrates three random forest classifiers to predict m6A modification sites. | / | http://www.cuilab.cn/sramp/ | [280] |
| m6AViewer | 2017 | It is a tools to detect, analyze and visualize the m6A peaks with sequencing dataset. | / | http://dna2.leeds.ac.uk/m6a | [281] |
| RNAMethPre | 2016 | It integrates multiple features of mRNA and provides a trained SVM classifier to predict m6A sites | Human/Mouse | | [282] |
| WHISTLE | 2019 | It is webserver for m6A site prediction and investigation of its potential biological process. | Human/Mouse/Yeast/ | http://whistle-epitranscriptome.com;www.xjtlu.edu.cn/biologicalsciences/whistle | [283] |
| AthMethPre | 2016 | It adopted SVM classifier to predict mRNA m6A modification sites. | Arabidopsis thaliana | http://bioinfo.tsinghua.edu.cn/AthMethPre/ | [284] |
| RFAthM6A | 2018 | It provides four independent models called RFPSNSP, RFPSDSP, RFKSNPF and RFKNF to predict m6A sites | Arabidopsis thaliana | https://github.com/nongdaxiaofeng/RFAthM6A | [285] |
| M6A-BiNP | 2021 | It provides two feature-encoding methods called PSP-PMI or PSP-PJMI to predict m6A sites | Human/Mouse/Rat | https://github.com/Mingzhao2017/M6A-BiNP | [286] |
| M6AMRFS | 2018 | It employs XGBoost classifier trained on dinucleotide binary encoding and Local position-specific dinucleotide frequency for m6A sites prediction | Human/Mouse | | [287] |
| DNN-m6A | 2021 | It provides DNN-based m6A prediction by adopting different strategies like TNC, ENAC and PSNP to extract sequence features and selecting feature subsets via elastic net | Human/Mouse/Rat | http://github.com/GD818/DNN-m6A | [288] |
| m6A-Maize | 2021 | It is a supervised learning model to predict m6A sites and provides a collection of traits-associated m6A mutations. | Maize | http://www.xjtlu.edu.cn/biologicalsciences/maize | [289] |
| TS-m6A-DL | 2021 | It is a predictor based on DNN trained by sequences with one-hot-encoding method | Human/Mouse/Rat | http://nsclbio.jbnu.ac.kr/tools/TS-m6A-DL/ | [290] |
| iRNA-m6A | 2020 | It adopted SVM classifier based on selected feature by mRMR to predict mRNA m6A modification sites. | Human/Mouse/Rat | http://lin-group.cn/server/iRNA-m6A | [291] |
| m6AGE | 2021 | It utilizes CatBoost model to predict m6A sites based on sequence-derived and graph embedding features | S. cerevisiae/ A.thaliana/ Human | https://github.com/bokunoBike/m6AGE | [292] |
| M6A-GSMS | 2021 | It employs GBDT to select features and train tracking model by combining seven basic classifiers. | A.thaliana/D.melanogaster/M.musculus/ S.cerevisiae/ Human | https://github.com/Wang-Jinyue/M6A-GSMS | [293] |
| iRNA-3typeA | 2018 | It is a SVM predictor based on PseKNC for m6A, m1A and A-to-I prediction. | / | http://lin-group.cn/server/iRNA-3typeA/ | [294] |
| iRNA-Methyl | 2015 | It is a predictor for m6A site based on sequence editing via pseudo nucleotide composition. | / | | [295] |
| BERMP | 2018 | It is a deep-learning classifier with bidirectional Gated Recurrent Unit (BGRU) for m6A prediction across multiple species. | Human/Mouse/Arabidopsis/Yeast | | [296] |
| M6APred-EL | 2018 | It is an ensemble of three SVM based on PS(k-mer) NP, PCPs and RFHC for m6A prediction. | / | | [297] |
| M6ATH | 2016 | It is a SVM-based predictor for identifying m6A sites in A.thaliana. | A.thaliana | | [298] |
| TargetM6A | 2016 | It is a SVM-based predictor trained by IFS-selected features that consist of PSNP/PSDP and NC transformed from RNA sequences. | / | http://csbio.njust.edu.cn/bioinf/TargetM6A | [299] |
| pRNAm-PC | 2016 | It is a predictor for m6A site based on sequence editing via pseudo nucleotide composition. | / | | [300] |
| M6A-HPCS | 2016 | It is an m6A site predictor based on HPCS algorithm under sequence feature representations including PseDNC, AC and CC. | S. cerevisiae (Example) | http://csbio.njust.edu.cn/bioinf/M6A-HPCS | [301] |
| RAM-ESVM | 2017 | It is an ensemble SVM classifiers trained by PseDNC, Motif features and gapped K-mers for m6A sites prediction in S. cerevisiae. | S. cerevisiae | | [302] |
| iRNA-PseColl | 2017 | It is a SVM-based platform to identify the occurrence sites for RNA modifications based on PseKNC. | / | | [303] |
| RAM-NPPS | 2017 | It is a SVM-based predictor for m6A identification rapidly based on a novel feature representation algorithm called multi-interval nucleotide pair position specificity. | Human/Arabidopsis/Yeast | | [304] |
| iMethyl-STTNC | 2018 | It is proposed for identification of m6A sites and is built on SVM, probabilistic neural network and KNN with feature extractions strategies, including PseDNC, PseTNC, STTNC and STNC. | | | [305] |
| Gene2Vec | 2019 | It is a soft vote predictor for m6A sites by integrating four different deep learning classifiers with shifting window. | Human/Mouse | | [306] |
| iRNA(m6A)-PseDNC | 2018 | It is a predictor for m6A site based on sequence editing via pseudo nucleotide composition. | S. cerevisiae | http://lin-group.cn/server/iRNA(m6A)-PseDNC.php | [307] |
| ConsRM | 2021 | It is a scoring framework that embedded with phastCons and phyloP to distinguish conserved m6A sites. | Human/Mouse/Fly/Zebrafish | https://www.xjtlu.edu.cn/biologicalsciences/con | [308] |
| Funm6AViewer | 2021 | It examines the distribution of m6A sites, prioritizes genes mediated by m6A modification and functionally gene regulatory network | / | https://www.xjtlu.edu.cn/biologicalsciences/funm6aviewer;https://github.com/NWPU-903PR/Funm6AViewer | [309] |
| xPore | 2021 | It identifies the m6A sites from nanopore direct RNA sequencing | Human/Mouse | https://github.com/GoekeLab/xpore | [310] |
| MASS | 2021 | It is a multi-task computational framework to capture m6A features and their regulatory functions across multiple species. | Human/Mouse/Yeast/ Arabidopsis/ Zebrafish/Rat/ Macaque/ Chimpanzee/ Fruit fly/Pig | https://github.com/mlcb-thu/MASS | [311] |
Methods detecting m6A sites
m6A modification sites can be detected directly by nanopore sequencing such as xPore and identified by isolating modified RNA fragments based on immunoprecipitation or specific RNA chemistry and combining them with sequencing [165, 166] (Table 11).
Table 11. Experimental methods for detecting m6A modification.
| Methods | Year | Characteristics | References |
| MeRIP-seq | 2015 | Combine m6A methylated mRNA fragments and high-throughput sequencing to detect m6A modification regions about 100nt-200nt. Cannot reflect the precise positions of m6A sites. | [167] |
| PA-m6A-seq | 2015 | Improve the resolution to 30nt around m6A modification sites via 4SU strengthening crosslinks. Can only detect m6A modification sites around 4SU operating sites with a resolution of 20nt-30nt. | [168] |
| miCLIP-seq | 2015 | Combine immunoprecipitation and high-throughput sequencing to detect m6A residues precisely at single-base resolution. | [169] |
| m6A-REF-seq | 2019 | Detect m6A sites in the whole transcriptome in an antibody-independent manner. | [170] |
| DART-seq | 2019 | Induce C-to-U deamination at sites adjacent to m6A residues to detect m6A. | [171] |
| m6A-label-seq | 2020 | Detect the m6A sites with a single-base resolution by marking the resources of methylation sites | [172] |
| m6A-SEAL | 2020 | Utilize streptavidin beads to enrich biotin-labeled m6A-modified sites for sequencing analysis. | [173] |
| m6A-seq2 | 2021 | Enhance robust m6A quantification at the site, gene, and sample resolution via multiple m6A immunoprecipitation techniques with bar code and mixed samples. | [174] |
| miCLIP2 | 2021 | Use low-input material to construct high-complexity libraries. Identify reliable m6A sites regardless of false positives induced by broad antibody reactivity, in combination with machine learning. | [175] |
| M6A-LAIC-seq | 2016 | Quantitatively detect m6A presence for genes or isoforms on a transcriptome-wide level based on the spike-in External RNA Controls Consortium. | [176] |
| SCARLET | 2014 | A digestion-based method to determine the percentage of m6A with single-nucleotide resolution for mRNAs and lncRNAs. It is not yet feasible for high-throughput applications. | [178] |
| SELECT | 2018 | Precisely distinguish m6A modification residues at a single base resolution. Does not require antibody IP or isotope labeling, thus reducing cost and can be widely used. | [177] |
| Nanopore direct-RNA sequencing | 2021 | Nanocompore can detect different RNA modifications with position accuracy in vitro. | [312] |
| HPLC-coupled mass spectrometry | 2014 | Short analysis time, good resolution and high sensitivity. | [313, 180] |
| Colorimetry | 2016 | Easily detect the overall level of RNA m6A modification. | [314] |
| Dot blot | 2017 | Dot blot analysis of RNA m6A is relatively easy, rapid and cost-effective. | [71] |
| PAR-CLIP | 2010 | It is based on incorporation of photoactivatable nucleoside analogs into nascent RNA. | [181] |
| Electrochemical immunosensor | 2015 | It showed a linear range for methylated RNA from 0.01 to 10 nM and the detection limit was 2.57 pM. | [182] |
A developed method called methylated RNA immunoprecipitation sequencing (MeRIP-seq) can combine m6A methylated mRNA fragments and high-throughput sequencing to detect m6A modification regions at about 100nt-200nt [167]. However, this method is complicated due to the largely overlapped reads for the similar m6A-containing fragments, thereby leading to prominent peaks spanning m6A residues, which indicates that MeRIP-seq method needs to reflect the precise positions of m6A sites. Then, one technique called PA-m6A-seq can improve the resolution to 30nt around m6A modification sites via 4SU strengthening crosslinks. However, PA-m6A-seq can only detect m6A modification sites around 4SU operating sites with a resolution of 20nt-30nt [168]. Another technology called miCLIP (m6A individual-nucleotide-resolution cross-linking and immunoprecipitation) can detect m6A residues precisely and overcome the disadvantages of PA-m6A-seq. Additionally, miCLIP-seq combines immunoprecipitation and high-throughput sequencing to identify m6A residues at high resolution, leading to the detection of m6A on snoRNA with high probability [169]. While m6A-REF-seq can quantify the m6A levels at single-base resolution and accurately detect m6A sites in the whole transcriptome in an antibody-independent manner, which is achieved by newly discovered RNA endonuclease MazF that is highly sensitive to m6A [170]. Another antibody-free method called DART-seq utilizes the fusion constructed by APOBEC1 (cytidine deaminase) and the YTH domain to induce C-to-U deamination at sites adjacent to m6A residues, thereby detecting m6A sites [171]. A recently developed method, termed m6A-label-seq, directly detects the m6A sites with a single-base resolution by marking the resources of methylation sites [172]. Briefly, METTL3/METTL14 utilize allyl-SAM or allyl-SeAM to install the methylation on RNA sites to obtain a6A, which can induce cyc-A formation under mild iodine conditions; hence m6A sites are identified via the mutations resulting from base mismatch caused by reverse transcribing from cyc-A to cDNA. At the same period, another technology named m6A-SEAL enables m6A sites to label biotin by methanethiosulfonate reacting with free sulfhydryl group on dm6A generated via m6A/hm6A/dm6A steps and utilizes streptavidin beads to enrich biotin-labeled m6A-modified sites for further sequencing analysis [173]. m6A-seq2, a newly updated method from m6A-seq, enhances robust m6A quantification at the site, gene, and sample resolution via multiple m6A immunoprecipitation techniques with bar codes and mixed samples [174]. Another updated technology, miCLIP2, characteristics of high-complexity libraries from less input material, identifies reliable m6A sites regardless of false positives induced by broad antibody reactivity, combined with machine learning [175].
The m6A-LAIC-seq method can quantify m6A presence for genes or isoforms on a transcriptome-wide level based on the spike-in External RNA Controls Consortium [176]. SELECT [177], MeRIP-qPCR [167] and MazF-qPCR [170] are often used to detect and quantify the m6A-containing fragments or genes. Method SCARLET can precisely distinguish m6A modification residues at single base resolution for mRNAs and lncRNAs based on specific RNase H, radiolabeling, and TLC [178]. And SCARLET can also be applied to detect m1A and Nm RNA modification sites. However, SCARLET is challenging to being widely used due to its complicated step and a significant dose of a radioactive isotope. And SCARLET can also be applied to detect m1A and Nm RNA modification sites. Single-base elongation and ligation-based qPCR amplification method (SELECT) is used for detecting m6A single base resolution in low abundance transcripts [177]. SELECT is based on two characteristics of m6A: (i) it blocks the single base extension of DNA polymerase during the reverse transcription process; (ii) reduces the nick ligation efficiency of ligases. This method does not require antibody IP or isotope labeling, thus reducing cost, and it can be widely used. SELECT enables the quantitative detection of m6A by qPCR. Recently, glyoxal and nitrite-mediated deamination of unmethylated adenosine (GLORI) realized the high efficiency and unbiased detection of single base m6A site and the absolute quantification of the modification level of m6A [179]. It is conceptually similar to bisulfite-sequencing-based quantification of DNA 5-methylcytosine, in which inosine is formed by efficient deamination of unmethylated adenosine using a catalytic system of glyoxal and nitrite. Inosine is paired with cytidine in the reverse transcription process and then read as guanosine (G) in the sequencing process, resulting in an A-to-G transformation. m6A remained the same and still read as A after sequencing. GLORI realized the absolute quantification of single base m6A by detecting the proportion of A in the sequence of the read segment. SELECT [177], MeRIP-qPCR [167], and MazF-qPCR [170] are often used to detect and quantify the m6A level of m6A-containing fragments or genes.
In addition, Mass spectrometry or colorimetry detects the global m6A level of RNAs, whereas high-performance liquid chromatography can identify specific m6A residues [180]. Additionally, dot blot technology is often used to observe m6A changes whereas it cannot be utilized to quantify m6A and identify the exact m6A modification sites [71]. However, researchers can use other methods, such as RNA photo-cross and immunoprecipitation (PAR-CLIP) [181] and electrochemical immunosensor [182] to detect m6A instead of the precise m6A modification sites.
Discussion and conclusion
In this review, we summarized the physiological functions of m6A regulators, the m6A-related databases, existing predictors for m6A sites, methods for detecting m6A, and the interplays of m6A and mRNA as well as ncRNAs in cancers. Generally, m6A writers install methyl groups on the 6th N of RNA adenosine whereas erasers remove the methylation. The readers recognize m6A sites of RNAs to regulate the translocation, splicing, degradation, stabilization, translation of RNAs, and other diverse biological functions. Interestingly, m6A may play oncogenic or suppressive roles in cancer initiation, progression, or resistance to therapy. How m6A affects cancer progression by regulating target genes depends on three factors: (1) whether the target gene acts as a tumor promoter or a tumor suppressor; (2) abnormal levels of m6A in cancer (depending on changes in writer or erase expression or activity); (3) regulation of target mRNA after methylation (as determined by reader). For instance, m6A on oncogenes can maintain their stabilities, MALAT1 in breast cancer or promote their degradation, XIST in CRC, as shown in Table 5. Tissue types play a more significant role in cancer genetics than previously realized, which should deserve consideration when designing treatments designed to inhibit cell proliferation. Well-designed bioinformatics analysis and mathematic modeling might be the key to elucidating the functional duality of m6A and maintaining physiological balance in different cellular contexts. The discrepancy among different research teams suggests that subsequent study requires shifting to a larger, multicenter paradigm to lay the foundation for the precision treatment of human tumors.
As emphasized above, m6A occurs on mRNA and ncRNAs and impacts the function of ncRNAs in cancers. For miRNAs, m6A affects the biogenesis and binding capacity with DGCR8 to participate in cancer progression. Concerning lncRNAs, m6A can function on lncRNAs to regulate their translocation, stabilization, and degradation, thereby leading to aberrant expression in cancers. Moreover, m6A acts on circRNA just like lncRNAs, except that m6A can also regulate the translation of circRNAs with coding ability. Conversely, lncRNAs and circRNAs can affect m6A levels directly by interacting with m6A regulators and indirectly by affecting m6A regulators through miRNAs, indicating that miRNAs can directly regulate the m6A by functioning on m6A regulators in cancers. This all suggests that m6A methylation on mRNAs or ncRNAs can be a potential target for cancer diagnosis, prognosis, and therapy. However, the investigation of the role of m6A in ncRNA remains lacking and needs further studies.
m6A regulators play controversially dual roles as oncogene or tumor suppressor genes across diverse cancers, which needs further exploration. And although accumulating evidence supports that m6A, mRNA, or ncRNA may be potential targets for cancer therapy, very few of them accomplish the clinical research stages, indicating a long way to go for clinical verification. In addition, there may be other RNA type-specific writers, readers, and erasers that have yet to be discovered, which will be crucial for gaining a better understanding of the function of m6A modification.
The rapid development of m6A detecting methods, m6A predicting methods trained by existing m6A modified sequences, and m6A-related databases significantly advance m6A investigation. Generally, Antibody-based m6A sequencing methods can preenrich for m6A signal and facilitate detection of low abundance RNA transcripts, but m6A-specific antibodies often have inherent bias and can also recognize other secondary modifications. On the contrary, chemical marker-based m6A sequencing technologies, such as m6A-SEAL and m6A-lable-seq can circumvent this problem. Endonuclease-based sequencing technologies such as MAZTER-seq and m6A-REF-seq utilize an endonuclease to detect m6A modification. However, due to the sequence bias of MazF, methods based on this enzyme can only detect part of the m6A site. Therefore, it is necessary to use these methods comprehensively in order to better detect m6A.
However, the explosion of multiple technologies has also increased confusion for researchers who lack explicit evaluations. Moreover, while mapping m6A was a critical advance, it required information on stoichiometry. New methods such as Mazter-Seq can accurately determine the stoichiometry of m6A at specific sites of particular mRNAs to determine if the transcriptome is truly dynamic and variable in different cellular and tumor progression contexts. Future studies should reveal to what extent m6A can dynamically regulate the epigenetic transcriptome of the tumor microenvironment and tumor-related immune environment to affect gene expression.
Author Contributions
Jun Chen supervised the project. Qingren Meng and Qian Zhou conceived and drafted the manuscript. Jun Chen and Heide Schatten edited the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest related to this study.
Funding
Supported by the funds for the construction of key medical disciplines in Shenzhen (No. SZXK076).
References
-
1.
Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021; 21:22–36. https://doi.org/10.1038/s41568-020-00306-0 [PubMed]
-
2.
Roundtree IA, He C. RNA epigenetics--chemical messages for posttranscriptional gene regulation. Curr Opin Chem Biol. 2016; 30:46–51. https://doi.org/10.1016/j.cbpa.2015.10.024 [PubMed]
-
3.
Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020; 20:303–22. https://doi.org/10.1038/s41568-020-0253-2 [PubMed]
-
4.
Liang W, Lin Z, Du C, Qiu D, Zhang Q. mRNA modification orchestrates cancer stem cell fate decisions. Mol Cancer. 2020; 19:38. https://doi.org/10.1186/s12943-020-01166-w [PubMed]
-
5.
Gu J, Zhan Y, Zhuo L, Zhang Q, Li G, Li Q, Qi S, Zhu J, Lv Q, Shen Y, Guo Y, Liu S, Xie T, Sui X. Biological functions of m6A methyltransferases. Cell Biosci. 2021; 11:15. https://doi.org/10.1186/s13578-020-00513-0 [PubMed]
-
6.
Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, Zhang R. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020; 48:6251–64. https://doi.org/10.1093/nar/gkaa347 [PubMed]
-
7.
Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, Bryson KM, Shachar R, Liman GLS, Burkhart BW, Gamage ST, Nobe Y, Briney CA, et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020; 583:638–43. https://doi.org/10.1038/s41586-020-2418-2 [PubMed]
-
8.
Malbec L, Zhang T, Chen YS, Zhang Y, Sun BF, Shi BY, Zhao YL, Yang Y, Yang YG. Dynamic methylome of internal mRNA N7-methylguanosine and its regulatory role in translation. Cell Res. 2019; 29:927–41. https://doi.org/10.1038/s41422-019-0230-z [PubMed]
-
9.
Shi H, Chai P, Jia R, Fan X. Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol Cancer. 2020; 19:78. https://doi.org/10.1186/s12943-020-01194-6 [PubMed]
-
10.
Zhou H, Rauch S, Dai Q, Cui X, Zhang Z, Nachtergaele S, Sepich C, He C, Dickinson BC. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat Methods. 2019; 16:1281–8. https://doi.org/10.1038/s41592-019-0550-4 [PubMed]
-
11.
Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018; 28:616–24. https://doi.org/10.1038/s41422-018-0040-8 [PubMed]
-
12.
Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, Fox SD, Zengeya TT, Andresson T, et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell. 2018; 175:1872–86.e24. https://doi.org/10.1016/j.cell.2018.10.030 [PubMed]
-
13.
Pandolfini L, Barbieri I, Bannister AJ, Hendrick A, Andrews B, Webster N, Murat P, Mach P, Brandi R, Robson SC, Migliori V, Alendar A, d'Onofrio M, et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell. 2019; 74:1278–90.e9. https://doi.org/10.1016/j.molcel.2019.03.040 [PubMed]
-
14.
Haruehanroengra P, Zheng YY, Zhou Y, Huang Y, Sheng J. RNA modifications and cancer. RNA Biol. 2020; 17:1560–75. https://doi.org/10.1080/15476286.2020.1722449 [PubMed]
-
15.
Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019; 20:608–24. https://doi.org/10.1038/s41580-019-0168-5 [PubMed]
-
16.
Lence T, Paolantoni C, Worpenberg L, Roignant JY. Mechanistic insights into m6A RNA enzymes. Biochim Biophys Acta Gene Regul Mech. 2019; 1862:222–9. https://doi.org/10.1016/j.bbagrm.2018.10.014 [PubMed]
-
17.
Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019; 112:108613. https://doi.org/10.1016/j.biopha.2019.108613 [PubMed]
-
18.
Su R, Dong L, Li Y, Gao M, He PC, Liu W, Wei J, Zhao Z, Gao L, Han L, Deng X, Li C, Prince E, et al. METTL16 exerts an m6A-independent function to facilitate translation and tumorigenesis. Nat Cell Biol. 2022; 24:205–16. https://doi.org/10.1038/s41556-021-00835-2 [PubMed]
-
19.
Kan RL, Chen J, Sallam T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2022; 38:182–93. https://doi.org/10.1016/j.tig.2021.06.014 [PubMed]
-
20.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015; 161:1388–99. https://doi.org/10.1016/j.cell.2015.05.014 [PubMed]
-
21.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–89. https://doi.org/10.1038/cr.2014.3 [PubMed]
-
22.
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016; 537:369–73. https://doi.org/10.1038/nature19342 [PubMed]
-
23.
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, Wang F, Wang X, Shen B, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018; 4:10. https://doi.org/10.1038/s41421-018-0019-0 [PubMed]
-
24.
Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, Lu X, Xiao W, Yang YG, Liu F. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017; 549:273–6. https://doi.org/10.1038/nature23883 [PubMed]
-
25.
Shafik AM, Zhang F, Guo Z, Dai Q, Pajdzik K, Li Y, Kang Y, Yao B, Wu H, He C, Allen EG, Duan R, Jin P. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer's disease. Genome Biol. 2021; 22:17. https://doi.org/10.1186/s13059-020-02249-z [PubMed]
-
26.
Ruszkowska A. METTL16, Methyltransferase-Like Protein 16: Current Insights into Structure and Function. Int J Mol Sci. 2021; 22:2176. https://doi.org/10.3390/ijms22042176 [PubMed]
-
27.
Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, Gross SS, Elemento O, Debart F, et al. Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature. 2017; 541:371–5. https://doi.org/10.1038/nature21022 [PubMed]
-
28.
Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, Jia G, Chen J, He C. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell. 2018; 71:973–85.e5. https://doi.org/10.1016/j.molcel.2018.08.011 [PubMed]
-
29.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013; 49:18–29. https://doi.org/10.1016/j.molcel.2012.10.015 [PubMed]
-
30.
Zhu LY, Zhu YR, Dai DJ, Wang X, Jin HC. Epigenetic regulation of alternative splicing. Am J Cancer Res. 2018; 8:2346–58. [PubMed]
-
31.
Melstrom L, Chen J. RNA N6-methyladenosine modification in solid tumors: new therapeutic frontiers. Cancer Gene Ther. 2020; 27:625–33. https://doi.org/10.1038/s41417-020-0160-4 [PubMed]
-
32.
Zhao Y, Shi Y, Shen H, Xie W. m6A-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. 2020; 13:35. https://doi.org/10.1186/s13045-020-00872-8 [PubMed]
-
33.
Patil DP, Pickering BF, Jaffrey SR. Reading m6A in the Transcriptome: m6A-Binding Proteins. Trends Cell Biol. 2018; 28:113–27. https://doi.org/10.1016/j.tcb.2017.10.001 [PubMed]
-
34.
Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, He E, Shen B, He C. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017; 6:e31311. https://doi.org/10.7554/eLife.31311 [PubMed]
-
35.
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, Wang X, Ma HL, Huang CM, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016; 61:507–19. https://doi.org/10.1016/j.molcel.2016.01.012 [PubMed]
-
36.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–20. https://doi.org/10.1038/nature12730 [PubMed]
-
37.
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016; 7:12626. https://doi.org/10.1038/ncomms12626 [PubMed]
-
38.
Wu E, Vashisht AA, Chapat C, Flamand MN, Cohen E, Sarov M, Tabach Y, Sonenberg N, Wohlschlegel J, Duchaine TF. A continuum of mRNP complexes in embryonic microRNA-mediated silencing. Nucleic Acids Res. 2017; 45:2081–98. https://doi.org/10.1093/nar/gkw872 [PubMed]
-
39.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017; 27:315–28. https://doi.org/10.1038/cr.2017.15 [PubMed]
-
40.
Zaccara S, Jaffrey SR. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell. 2020; 181:1582–95.e18. https://doi.org/10.1016/j.cell.2020.05.012 [PubMed]
-
41.
Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, Liu J, Sun Z. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol Cancer. 2020; 19:105. https://doi.org/10.1186/s12943-020-01224-3 [PubMed]
-
42.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015; 162:1299–308. https://doi.org/10.1016/j.cell.2015.08.011 [PubMed]
-
43.
Piñol-Roma S, Dreyfuss G. Cell cycle-regulated phosphorylation of the pre-mRNA-binding (heterogeneous nuclear ribonucleoprotein) C proteins. Mol Cell Biol. 1993; 13:5762–70. https://doi.org/10.1128/mcb.13.9.5762-5770.1993 [PubMed]
-
44.
Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Ż, Pan JN, He C, Parisien M, Pan T. Regulation of Co-transcriptional Pre-mRNA Splicing by m6A through the Low-Complexity Protein hnRNPG. Mol Cell. 2019; 76:70–81.e9. https://doi.org/10.1016/j.molcel.2019.07.005 [PubMed]
-
45.
Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015; 163:999–1010. https://doi.org/10.1016/j.cell.2015.10.012 [PubMed]
-
46.
des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015; 525:491–5. https://doi.org/10.1038/nature14891 [PubMed]
-
47.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Hüttelmaier S, Skibbe JR, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018; 20:285–95. https://doi.org/10.1038/s41556-018-0045-z [PubMed]
-
48.
Chen B, Li Y, Song R, Xue C, Xu F. Functions of RNA N6-methyladenosine modification in cancer progression. Mol Biol Rep. 2019; 46:2567–75. https://doi.org/10.1007/s11033-019-04655-4 [PubMed]
-
49.
Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, Yuan W, Kan Q, Sun Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019; 12:121. https://doi.org/10.1186/s13045-019-0805-7 [PubMed]
-
50.
Pan Y, Ma P, Liu Y, Li W, Shu Y. Multiple functions of m6A RNA methylation in cancer. J Hematol Oncol. 2018; 11:48. https://doi.org/10.1186/s13045-018-0590-8 [PubMed]
-
51.
Liu S, Li G, Li Q, Zhang Q, Zhuo L, Chen X, Zhai B, Sui X, Chen K, Xie T. The roles and mechanisms of YTH domain-containing proteins in cancer development and progression. Am J Cancer Res. 2020; 10:1068–84. [PubMed]
-
52.
Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018; 37:522–33. https://doi.org/10.1038/onc.2017.351 [PubMed]
-
53.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, Zhou J, Sun B, Zou X, Wang S. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020; 69:1193–205. https://doi.org/10.1136/gutjnl-2019-319639 [PubMed]
-
54.
Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018; 67:2254–70. https://doi.org/10.1002/hep.29683 [PubMed]
-
55.
Liu L, Wang J, Sun G, Wu Q, Ma J, Zhang X, Huang N, Bian Z, Gu S, Xu M, Yin M, Sun F, Pan Q. m6A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol Cancer. 2019; 18:188. https://doi.org/10.1186/s12943-019-1119-7 [PubMed]
-
56.
Zhu W, Si Y, Xu J, Lin Y, Wang JZ, Cao M, Sun S, Ding Q, Zhu L, Wei JF. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med. 2020; 24:3521–33. https://doi.org/10.1111/jcmm.15042 [PubMed]
-
57.
Gao Q, Zheng J, Ni Z, Sun P, Yang C, Cheng M, Wu M, Zhang X, Yuan L, Zhang Y, Li Y. The m6A Methylation-Regulated AFF4 Promotes Self-Renewal of Bladder Cancer Stem Cells. Stem Cells Int. 2020; 2020:8849218. https://doi.org/10.1155/2020/8849218 [PubMed]
-
58.
Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, Liang Y, Zhu F, Zhang Y, Zhang X, Yuan Q, Li Y. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019; 38:3667–80. https://doi.org/10.1038/s41388-019-0683-z [PubMed]
-
59.
Wanna-Udom S, Terashima M, Lyu H, Ishimura A, Takino T, Sakari M, Tsukahara T, Suzuki T. The m6A methyltransferase METTL3 contributes to Transforming Growth Factor-beta-induced epithelial-mesenchymal transition of lung cancer cells through the regulation of JUNB. Biochem Biophys Res Commun. 2020; 524:150–5. https://doi.org/10.1016/j.bbrc.2020.01.042 [PubMed]
-
60.
Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, Santisteban P, George RE, Richards WG, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018; 561:556–60. https://doi.org/10.1038/s41586-018-0538-8 [PubMed]
-
61.
Zhou R, Gao Y, Lv D, Wang C, Wang D, Li Q. METTL3 mediated m6A modification plays an oncogenic role in cutaneous squamous cell carcinoma by regulating ΔNp63. Biochem Biophys Res Commun. 2019; 515:310–7. https://doi.org/10.1016/j.bbrc.2019.05.155 [PubMed]
-
62.
Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, Doki Y, Mori M, Ishii H, Ogawa K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018; 52:621–9. https://doi.org/10.3892/ijo.2017.4219 [PubMed]
-
63.
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, Riggs AD, He C, Shi Y. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017; 18:2622–34. https://doi.org/10.1016/j.celrep.2017.02.059 [PubMed]
-
64.
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, Li S, Tan L, Mai D, Li G, Pan L, Zheng Y, Su J, et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019; 10:1858. https://doi.org/10.1038/s41467-019-09712-x [PubMed]
-
65.
Yang F, Yuan WQ, Li J, Luo YQ. Knockdown of METTL14 suppresses the malignant progression of non-small cell lung cancer by reducing Twist expression. Oncol Lett. 2021; 22:847. https://doi.org/10.3892/ol.2021.13108 [PubMed]
-
66.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, Sheng Y, Wang Y, Wunderlich M, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell. 2018; 22:191–205.e9. https://doi.org/10.1016/j.stem.2017.11.016 [PubMed]
-
67.
Chen Y, Peng C, Chen J, Chen D, Yang B, He B, Hu W, Zhang Y, Liu H, Dai L, Xie H, Zhou L, Wu J, Zheng S. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019; 18:127. https://doi.org/10.1186/s12943-019-1053-8 [PubMed]
-
68.
Naren D, Yan T, Gong Y, Huang J, Zhang D, Sang L, Zheng X, Li Y. High Wilms' tumor 1 associating protein expression predicts poor prognosis in acute myeloid leukemia and regulates m6A methylation of MYC mRNA. J Cancer Res Clin Oncol. 2021; 147:33–47. https://doi.org/10.1007/s00432-020-03373-w [PubMed]
-
69.
Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia TS, Zhou WB, Wang S, Ding Q, Wei JF. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene. 2019; 38:6123–41. https://doi.org/10.1038/s41388-019-0861-z [PubMed]
-
70.
Lan T, Li H, Zhang D, Xu L, Liu H, Hao X, Yan X, Liao H, Chen X, Xie K, Li J, Liao M, Huang J, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019; 18:186. https://doi.org/10.1186/s12943-019-1106-z [PubMed]
-
71.
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell. 2017; 31:127–41. https://doi.org/10.1016/j.ccell.2016.11.017 [PubMed]
-
72.
Qing Y, Dong L, Gao L, Li C, Li Y, Han L, Prince E, Tan B, Deng X, Wetzel C, Shen C, Gao M, Chen Z, et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol Cell. 2021; 81:922–39.e9. https://doi.org/10.1016/j.molcel.2020.12.026 [PubMed]
-
73.
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, Wu X, Wan G. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019; 18:46. https://doi.org/10.1186/s12943-019-1004-4 [PubMed]
-
74.
Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, Aplin AE, Lu Z, Hwang S, He C, He YY. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019; 10:2782. https://doi.org/10.1038/s41467-019-10669-0 [PubMed]
-
75.
Liu Y, Liang G, Xu H, Dong W, Dong Z, Qiu Z, Zhang Z, Li F, Huang Y, Li Y, Wu J, Yin S, Zhang Y, et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 2021; 33:1221–33.e11. https://doi.org/10.1016/j.cmet.2021.04.001 [PubMed]
-
76.
Xiao L, Li X, Mu Z, Zhou J, Zhou P, Xie C, Jiang S. FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res. 2020; 80:3945–58. https://doi.org/10.1158/0008-5472.CAN-20-0132 [PubMed]
-
77.
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, Ni T, Zhang ZS, Zhang T, Li C, Han L, Zhu Z, Lian F, et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019; 35:677–91.e10. https://doi.org/10.1016/j.ccell.2019.03.006 [PubMed]
-
78.
Yang X, Shao F, Guo D, Wang W, Wang J, Zhu R, Gao Y, He J, Lu Z. WNT/β-catenin-suppressed FTO expression increases m6A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis. Cell Death Dis. 2021; 12:462. https://doi.org/10.1038/s41419-021-03739-z [PubMed]
-
79.
Rong ZX, Li Z, He JJ, Liu LY, Ren XX, Gao J, Mu Y, Guan YD, Duan YM, Zhang XP, Zhang DX, Li N, Deng YZ, Sun LQ. Downregulation of Fat Mass and Obesity Associated (FTO) Promotes the Progression of Intrahepatic Cholangiocarcinoma. Front Oncol. 2019; 9:369. https://doi.org/10.3389/fonc.2019.00369 [PubMed]
-
80.
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, Majumder S, He C, Huang S. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017; 31:591–606.e6. https://doi.org/10.1016/j.ccell.2017.02.013 [PubMed]
-
81.
Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, Chen H, Su R, Yin Z, Li W, Deng X, Chen Y, Hu YC, et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell. 2020; 27:64–80.e9. https://doi.org/10.1016/j.stem.2020.04.009 [PubMed]
-
82.
Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019; 38:163. https://doi.org/10.1186/s13046-019-1159-2 [PubMed]
-
83.
Chen Y, Zhao Y, Chen J, Peng C, Zhang Y, Tong R, Cheng Q, Yang B, Feng X, Lu Y, Xie H, Zhou L, Wu J, Zheng S. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m6A-guided epigenetic inhibition of LYPD1. Mol Cancer. 2020; 19:123. https://doi.org/10.1186/s12943-020-01239-w [PubMed]
-
84.
Liu X, Qin J, Gao T, Li C, He B, Pan B, Xu X, Chen X, Zeng K, Xu M, Zhu C, Pan Y, Sun H, et al. YTHDF1 Facilitates the Progression of Hepatocellular Carcinoma by Promoting FZD5 mRNA Translation in an m6A-Dependent Manner. Mol Ther Nucleic Acids. 2020; 22:750–65. https://doi.org/10.1016/j.omtn.2020.09.036 [PubMed]. Retraction in: Mol Ther Nucleic Acids. 2022; 28:571. https://doi.org/10.1016/j.omtn.2022.04.027 [PubMed]
-
85.
Anita R, Paramasivam A, Priyadharsini JV, Chitra S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res. 2020; 10:2546–54. [PubMed]
-
86.
Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, Shen Q, Xu P, Zeng L, Zhou Y, Huang Y, Yang Z, Zhou J, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019; 10:4892. https://doi.org/10.1038/s41467-019-12801-6 [PubMed]
-
87.
Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 Regulates Tumorigenicity and Cancer Stem Cell-Like Activity in Human Colorectal Carcinoma. Front Oncol. 2019; 9:332. https://doi.org/10.3389/fonc.2019.00332 [PubMed]
-
88.
Orouji E, Peitsch WK, Orouji A, Houben R, Utikal J. Oncogenic Role of an Epigenetic Reader of m6A RNA Modification: YTHDF1 in Merkel Cell Carcinoma. Cancers (Basel). 2020; 12:202. https://doi.org/10.3390/cancers12010202 [PubMed]
-
89.
Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai D, Kudo T, Hata T, Matsuda C, et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2017; 9:7476–86. https://doi.org/10.18632/oncotarget.23554 [PubMed]
-
90.
Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, Huang X, Liu Y, Wang J, Dougherty U, Bissonnette MB, Shen B, Weichselbaum RR, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019; 566:270–4. https://doi.org/10.1038/s41586-019-0916-x [PubMed]
-
91.
Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, Ge S, Jia R, Yang YG, Fan X. m6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer. 2019; 18:161. https://doi.org/10.1186/s12943-019-1088-x [PubMed]
-
92.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA, Sepulveda C, Vukovic M, Allen L, Sarapuu A, et al. Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019; 25:137–48.e6. https://doi.org/10.1016/j.stem.2019.03.021 [PubMed]
-
93.
Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, Lu Y, Zeng J, Du F, Gong A, Xu M. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017; 16:2259–71. https://doi.org/10.1080/15384101.2017.1380125 [PubMed]
-
94.
Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, Ma H, Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019; 442:252–61. https://doi.org/10.1016/j.canlet.2018.11.006 [PubMed]
-
95.
Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, Hu B, Zhou J, Zhao Z, Feng M, Zhang H, Shen B, Huang X, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019; 18:163. https://doi.org/10.1186/s12943-019-1082-3 [PubMed]
-
96.
Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, Cao X. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc Natl Acad Sci U S A. 2019; 116:976–81. https://doi.org/10.1073/pnas.1812536116 [PubMed]
-
97.
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019; 18:143. https://doi.org/10.1186/s12943-019-1079-y [PubMed]
-
98.
Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X, Ren F, Cui G, Sun R. Expression patterns and prognostic value of m6A-related genes in colorectal cancer. Am J Transl Res. 2019; 11:3972–91. [PubMed]
-
99.
Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, Mori M, Sahara H. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016; 376:34–42. https://doi.org/10.1016/j.canlet.2016.02.022 [PubMed]
-
100.
Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013; 70:2657–75. https://doi.org/10.1007/s00018-012-1186-z [PubMed]
-
101.
Zhang L, Wan Y, Zhang Z, Jiang Y, Gu Z, Ma X, Nie S, Yang J, Lang J, Cheng W, Zhu L. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics. 2021; 11:1100–14. https://doi.org/10.7150/thno.49345 [PubMed]
-
102.
He RZ, Jiang J, Luo DX. M6A modification of circNSUN2 promotes colorectal liver metastasis. Genes Dis. 2019; 8:6–7. https://doi.org/10.1016/j.gendis.2019.12.002 [PubMed]
-
103.
Huang H, Weng H, Chen J. m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020; 37:270–88. https://doi.org/10.1016/j.ccell.2020.02.004 [PubMed]
-
104.
Li H, Wu H, Wang Q, Ning S, Xu S, Pang D. Dual effects of N6-methyladenosine on cancer progression and immunotherapy. Mol Ther Nucleic Acids. 2021; 24:25–39. https://doi.org/10.1016/j.omtn.2021.02.001 [PubMed]
-
105.
Erson-Bensan AE, Begik O. m6A Modification and Implications for microRNAs. Microrna. 2017; 6:97–101. https://doi.org/10.2174/2211536606666170511102219 [PubMed]
-
106.
Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014; 15:313–26. https://doi.org/10.1038/nrm3785 [PubMed]
-
107.
Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015; 519:482–5. https://doi.org/10.1038/nature14281 [PubMed]
-
108.
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, Wei JF, Yang H. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019; 18:110. https://doi.org/10.1186/s12943-019-1036-9 [PubMed]
-
109.
Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019; 38:393. https://doi.org/10.1186/s13046-019-1408-4 [PubMed]
-
110.
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, Dinglin X, Ma S, Li D, Wu Y, Peng Y, Huang H, Chen L. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019; 18:181. https://doi.org/10.1186/s12943-019-1108-x [PubMed]
-
111.
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017; 65:529–43. https://doi.org/10.1002/hep.28885 [PubMed]
-
112.
Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, Pan B, He B, Pan Y, Sun H, Xia X, Wang S. RETRACTED: METTL14 Suppresses CRC Progression via Regulating N6-Methyladenosine-Dependent Primary miR-375 Processing. Mol Ther. 2020; 28:599–612. https://doi.org/10.1016/j.ymthe.2019.11.016 [PubMed]. Retraction in: Mol Ther. 2022; 30:2640. https://doi.org/10.1016/j.ymthe.2022.03.017 [PubMed]
-
113.
Park YM, Hwang SJ, Masuda K, Choi KM, Jeong MR, Nam DH, Gorospe M, Kim HH. Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4. Mol Cell Biol. 2012; 32:4237–44. https://doi.org/10.1128/MCB.00443-12 [PubMed]
-
114.
Shah A, Rashid F, Awan HM, Hu S, Wang X, Chen L, Shan G. The DEAD-Box RNA Helicase DDX3 Interacts with m6A RNA Demethylase ALKBH5. Stem Cells Int. 2017; 2017:8596135. https://doi.org/10.1155/2017/8596135 [PubMed]
-
115.
Han X, Guo J, Fan Z. Interactions between m6A modification and miRNAs in malignant tumors. Cell Death Dis. 2021; 12:598. https://doi.org/10.1038/s41419-021-03868-5 [PubMed]
-
116.
Ye S, Song W, Xu X, Zhao X, Yang L. IGF2BP2 promotes colorectal cancer cell proliferation and survival through interfering with RAF-1 degradation by miR-195. FEBS Lett. 2016; 590:1641–50. https://doi.org/10.1002/1873-3468.12205 [PubMed]
-
117.
Zhou Y, Huang T, Siu HL, Wong CC, Dong Y, Wu F, Zhang B, Wu WK, Cheng AS, Yu J, To KF, Kang W. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer. 2017; 16:77. https://doi.org/10.1186/s12943-017-0647-2 [PubMed]
-
118.
Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, Liu Y, Ye L, Li Y, Zhang X. MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3'-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem. 2017; 292:3614–23. https://doi.org/10.1074/jbc.M116.749689 [PubMed]
-
119.
Dai F, Wu Y, Lu Y, An C, Zheng X, Dai L, Guo Y, Zhang L, Li H, Xu W, Gao W. Crosstalk between RNA m6A Modification and Non-coding RNA Contributes to Cancer Growth and Progression. Mol Ther Nucleic Acids. 2020; 22:62–71. https://doi.org/10.1016/j.omtn.2020.08.004 [PubMed]
-
120.
He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019; 18:176. https://doi.org/10.1186/s12943-019-1109-9 [PubMed]
-
121.
He RZ, Jiang J, Luo DX. The functions of N6-methyladenosine modification in lncRNAs. Genes Dis. 2020; 7:598–605. https://doi.org/10.1016/j.gendis.2020.03.005 [PubMed]
-
122.
Dinescu S, Ignat S, Lazar AD, Constantin C, Neagu M, Costache M. Epitranscriptomic Signatures in lncRNAs and Their Possible Roles in Cancer. Genes (Basel). 2019; 10:52. https://doi.org/10.3390/genes10010052 [PubMed]
-
123.
Yang X, Zhang S, He C, Xue P, Zhang L, He Z, Zang L, Feng B, Sun J, Zheng M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020; 19:46. https://doi.org/10.1186/s12943-020-1146-4 [PubMed]
-
124.
Zhao C, Ling X, Xia Y, Yan B, Guan Q. The m6A methyltransferase METTL3 controls epithelial-mesenchymal transition, migration and invasion of breast cancer through the MALAT1/miR-26b/HMGA2 axis. Cancer Cell Int. 2021; 21:441. https://doi.org/10.1186/s12935-021-02113-5 [PubMed]
-
125.
Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, Cao M, Cai J, Wu J, Wang X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020; 13:5. https://doi.org/10.1186/s13045-019-0839-x [PubMed]
-
126.
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, Huang XD, Liu RQ, Chen F, He XJ, Kou J, Zhang J, Wen X, et al. Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 2019; 79:4612–26. https://doi.org/10.1158/0008-5472.CAN-19-0799 [PubMed]
-
127.
Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, Li J, An P, Lu L, Luo N, Du J, Shan H, Liu H, Wang H. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019; 18:87. https://doi.org/10.1186/s12943-019-1014-2 [PubMed]
-
128.
Xue L, Li J, Lin Y, Liu D, Yang Q, Jian J, Peng J. m6 A transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer. J Cell Physiol. 2021; 236:2649–58. https://doi.org/10.1002/jcp.30023 [PubMed]
-
129.
Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, Yang D, Zheng ZC, Zhao Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019; 75:379–89. https://doi.org/10.1007/s13105-019-00690-8 [PubMed]
-
130.
Chen S, Zhou L, Wang Y. ALKBH5-mediated m6A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020; 20:34. https://doi.org/10.1186/s12935-020-1105-6 [PubMed]
-
131.
He Y, Yue H, Cheng Y, Ding Z, Xu Z, Lv C, Wang Z, Wang J, Yin C, Hao H, Chen C. ALKBH5-mediated m6A demethylation of KCNK15-AS1 inhibits pancreatic cancer progression via regulating KCNK15 and PTEN/AKT signaling. Cell Death Dis. 2021; 12:1121. https://doi.org/10.1038/s41419-021-04401-4 [PubMed]
-
132.
Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, Ma Y, Fang J, Wang Y, Cao W, Guan F. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021; 40:294. https://doi.org/10.1186/s13046-021-02096-1 [PubMed]
-
133.
Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, Mo YY, Yang L. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020; 27:1782–94. https://doi.org/10.1038/s41418-019-0461-z [PubMed]
-
134.
Ye M, Dong S, Hou H, Zhang T, Shen M. Oncogenic Role of Long Noncoding RNAMALAT1 in Thyroid Cancer Progression through Regulation of the miR-204/IGF2BP2/m6A-MYC Signaling. Mol Ther Nucleic Acids. 2020; 23:1–12. https://doi.org/10.1016/j.omtn.2020.09.023 [PubMed]
-
135.
Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX, Liu J, Luo XJ, Meng Q, Pu HY, Wang YN, Hu PS, Liu ZX, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019; 18:174. https://doi.org/10.1186/s12943-019-1105-0 [PubMed]
-
136.
Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W, Huang Z. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020; 521:887–93. https://doi.org/10.1016/j.bbrc.2019.11.016 [PubMed]
-
137.
Zhu P, He F, Hou Y, Tu G, Li Q, Jin T, Zeng H, Qin Y, Wan X, Qiao Y, Qiu Y, Teng Y, Liu M. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021; 40:1609–27. https://doi.org/10.1038/s41388-020-01638-9 [PubMed]
-
138.
Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, Xu W, Jiang T, Feng L, Shin VY, Wang X, Jin H. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019; 10:383. https://doi.org/10.1038/s41419-019-1585-2 [PubMed]
-
139.
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015; 12:381–8. https://doi.org/10.1080/15476286.2015.1020271 [PubMed]
-
140.
Wang X, Ma R, Zhang X, Cui L, Ding Y, Shi W, Guo C, Shi Y. Crosstalk between N6-methyladenosine modification and circular RNAs: current understanding and future directions. Mol Cancer. 2021; 20:121. https://doi.org/10.1186/s12943-021-01415-6 [PubMed]
-
141.
Meng Y, Hao D, Huang Y, Jia S, Zhang J, He X, Liu D, Sun L. Circular RNA circNRIP1 plays oncogenic roles in the progression of osteosarcoma. Mamm Genome. 2021; 32:448–56. https://doi.org/10.1007/s00335-021-09891-3 [PubMed]
-
142.
Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, Shao B, Sun Z, Ji Z. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021; 20:93. https://doi.org/10.1186/s12943-021-01372-0 [PubMed]
-
143.
Li Z, Yang HY, Dai XY, Zhang X, Huang YZ, Shi L, Wei JF, Ding Q. CircMETTL3, upregulated in a m6A-dependent manner, promotes breast cancer progression. Int J Biol Sci. 2021; 17:1178–90. https://doi.org/10.7150/ijbs.57783 [PubMed]
-
144.
Ji F, Lu Y, Chen S, Yu Y, Lin X, Zhu Y, Luo X. IGF2BP2-modified circular RNA circARHGAP12 promotes cervical cancer progression by interacting m6A/FOXM1 manner. Cell Death Discov. 2021; 7:215. https://doi.org/10.1038/s41420-021-00595-w [PubMed]
-
145.
Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, Zhan X, Laimins L, Wang RC. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019; 10:2300. https://doi.org/10.1038/s41467-019-10246-5 [PubMed]
-
146.
Chen C, Yuan W, Zhou Q, Shao B, Guo Y, Wang W, Yang S, Guo Y, Zhao L, Dang Q, Yang X, Wang G, Kang Q, et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics. 2021; 11:4298–315. https://doi.org/10.7150/thno.51342 [PubMed]
-
147.
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, Wang F, Ma NF, Guan X, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019; 10:4695. https://doi.org/10.1038/s41467-019-12651-2 [PubMed]
-
148.
Rong D, Wu F, Lu C, Sun G, Shi X, Chen X, Dai Y, Zhong W, Hao X, Zhou J, Xia Y, Tang W, Wang X. m6A modification of circHPS5 and hepatocellular carcinoma progression through HMGA2 expression. Mol Ther Nucleic Acids. 2021; 26:637–48. https://doi.org/10.1016/j.omtn.2021.09.001 [PubMed]
-
149.
Rao X, Lai L, Li X, Wang L, Li A, Yang Q. N6 -methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life. 2021; 73:408–17. https://doi.org/10.1002/iub.2438 [PubMed]
-
150.
Liu Z, Wang T, She Y, Wu K, Gu S, Li L, Dong C, Chen C, Zhou Y. N6-methyladenosine-modified circIGF2BP3 inhibits CD8+ T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021; 20:105. https://doi.org/10.1186/s12943-021-01398-4 [PubMed]
-
151.
Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou J, Jin H, Zhao A, Tang WW, Cao XF. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018; 8:10111. https://doi.org/10.1038/s41598-018-27837-9 [PubMed]
-
152.
Shen Y, Li C, Zhou L, Huang JA. G protein-coupled oestrogen receptor promotes cell growth of non-small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A methylated NOTCH1 signalling. J Cell Mol Med. 2021; 25:284–96. https://doi.org/10.1111/jcmm.15997 [PubMed]
-
153.
Zhang Z, Zhu H, Hu J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 2021; 12:219. https://doi.org/10.1038/s41419-021-03486-1 [PubMed]
-
154.
Wei W, Sun J, Zhang H, Xiao X, Huang C, Wang L, Zhong H, Jiang Y, Zhang X, Jiang G. Circ0008399 Interaction with WTAP Promotes Assembly and Activity of the m6A Methyltransferase Complex and Promotes Cisplatin Resistance in Bladder Cancer. Cancer Res. 2021; 81:6142–56. https://doi.org/10.1158/0008-5472.CAN-21-1518 [PubMed]
-
155.
Chi F, Cao Y, Chen Y. Analysis and Validation of circRNA-miRNA Network in Regulating m6A RNA Methylation Modulators Reveals CircMAP2K4/miR-139-5p/YTHDF1 Axis Involving the Proliferation of Hepatocellular Carcinoma. Front Oncol. 2021; 11:560506. https://doi.org/10.3389/fonc.2021.560506 [PubMed]
-
156.
Mo WL, Deng LJ, Cheng Y, Yu WJ, Yang YH, Gu WD. Circular RNA hsa_circ_0072309 promotes tumorigenesis and invasion by regulating the miR-607/FTO axis in non-small cell lung carcinoma. Aging (Albany NY). 2021; 13:11629–45. https://doi.org/10.18632/aging.202856 [PubMed]
-
157.
Tang Y, Chen K, Song B, Ma J, Wu X, Xu Q, Wei Z, Su J, Liu G, Rong R, Lu Z, de Magalhães JP, Rigden DJ, Meng J. m6A-Atlas: a comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res. 2021; 49:D134–43. https://doi.org/10.1093/nar/gkaa692 [PubMed]
-
158.
Deng S, Zhang H, Zhu K, Li X, Ye Y, Li R, Liu X, Lin D, Zuo Z, Zheng J. M6A2Target: a comprehensive database for targets of m6A writers, erasers and readers. Brief Bioinform. 2021; 22:bbaa055. https://doi.org/10.1093/bib/bbaa055 [PubMed]
-
159.
Luo X, Li H, Liang J, Zhao Q, Xie Y, Ren J, Zuo Z. RMVar: an updated database of functional variants involved in RNA modifications. Nucleic Acids Res. 2021; 49:D1405–12. https://doi.org/10.1093/nar/gkaa811 [PubMed]
-
160.
Chen K, Song B, Tang Y, Wei Z, Xu Q, Su J, de Magalhães JP, Rigden DJ, Meng J. RMDisease: a database of genetic variants that affect RNA modifications, with implications for epitranscriptome pathogenesis. Nucleic Acids Res. 2021; 49:D1396–404. https://doi.org/10.1093/nar/gkaa790 [PubMed]
-
161.
Liu H, Wang H, Wei Z, Zhang S, Hua G, Zhang SW, Zhang L, Gao SJ, Meng J, Chen X, Huang Y. MeT-DB V2.0: elucidating context-specific functions of N6-methyl-adenosine methyltranscriptome. Nucleic Acids Res. 2018; 46:D281–7. https://doi.org/10.1093/nar/gkx1080 [PubMed]
-
162.
Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S, Zheng LL, Qu LH, Yang JH. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018; 46:D327–34. https://doi.org/10.1093/nar/gkx934 [PubMed]
-
163.
Liu S, Zhu A, He C, Chen M. REPIC: a database for exploring the N6-methyladenosine methylome. Genome Biol. 2020; 21:100. https://doi.org/10.1186/s13059-020-02012-4 [PubMed]
-
164.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–6. https://doi.org/10.1038/nature11112 [PubMed]
-
165.
Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, Hall AJ, Barton GJ, Simpson GG. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. Elife. 2020; 9:e49658. https://doi.org/10.7554/eLife.49658 [PubMed]
-
166.
Gao Y, Liu X, Wu B, Wang H, Xi F, Kohnen MV, Reddy ASN, Gu L. Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing. Genome Biol. 2021; 22:22. https://doi.org/10.1186/s13059-020-02241-7 [PubMed]
-
167.
Dominissini D, Moshitch-Moshkovitz S, Amariglio N, Rechavi G. Transcriptome-Wide Mapping of N6-Methyladenosine by m6A-Seq. Methods Enzymol. 2015; 560:131–47. https://doi.org/10.1016/bs.mie.2015.03.001 [PubMed]
-
168.
Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N, Han D, Dominissini D, Dai Q, Pan T, He C. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew Chem Int Ed Engl. 2015; 54:1587–90. https://doi.org/10.1002/anie.201410647 [PubMed]
-
169.
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015; 12:767–72. https://doi.org/10.1038/nmeth.3453 [PubMed]
-
170.
Zhang Z, Chen LQ, Zhao YL, Yang CG, Roundtree IA, Zhang Z, Ren J, Xie W, He C, Luo GZ. Single-base mapping of m6A by an antibody-independent method. Sci Adv. 2019; 5:eaax0250. https://doi.org/10.1126/sciadv.aax0250 [PubMed]
-
171.
Meyer KD. DART-seq: an antibody-free method for global m6A detection. Nat Methods. 2019; 16:1275–80. https://doi.org/10.1038/s41592-019-0570-0 [PubMed]
-
172.
Shu X, Cao J, Cheng M, Xiang S, Gao M, Li T, Ying X, Wang F, Yue Y, Lu Z, Dai Q, Cui X, Ma L, et al. A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat Chem Biol. 2020; 16:887–95. https://doi.org/10.1038/s41589-020-0526-9 [PubMed]
-
173.
Wang Y, Xiao Y, Dong S, Yu Q, Jia G. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat Chem Biol. 2020; 16:896–903. https://doi.org/10.1038/s41589-020-0525-x [PubMed]
-
174.
Dierks D, Garcia-Campos MA, Uzonyi A, Safra M, Edelheit S, Rossi A, Sideri T, Varier RA, Brandis A, Stelzer Y, van Werven F, Scherz-Shouval R, Schwartz S. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat Methods. 2021; 18:1060–7. https://doi.org/10.1038/s41592-021-01242-z [PubMed]
-
175.
Körtel N, Rücklé C, Zhou Y, Busch A, Hoch-Kraft P, Sutandy FXR, Haase J, Pradhan M, Musheev M, Ostareck D, Ostareck-Lederer A, Dieterich C, Hüttelmaier S, et al. Deep and accurate detection of m6A RNA modifications using miCLIP2 and m6Aboost machine learning. Nucleic Acids Res. 2021; 49:e92. https://doi.org/10.1093/nar/gkab485 [PubMed]
-
176.
Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, Xing Y, Giallourakis CC. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods. 2016; 13:692–8. https://doi.org/10.1038/nmeth.3898 [PubMed]
-
177.
Xiao Y, Wang Y, Tang Q, Wei L, Zhang X, Jia G. An Elongation- and Ligation-Based qPCR Amplification Method for the Radiolabeling-Free Detection of Locus-Specific N6 -Methyladenosine Modification. Angew Chem Int Ed Engl. 2018; 57:15995–6000. https://doi.org/10.1002/anie.201807942 [PubMed]
-
178.
Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013; 19:1848–56. https://doi.org/10.1261/rna.041178.113 [PubMed]
-
179.
Liu C, Sun H, Yi Y, Shen W, Li K, Xiao Y, Li F, Li Y, Hou Y, Lu B, Liu W, Meng H, Peng J, et al. Absolute quantification of single-base m6A methylation in the mammalian transcriptome using GLORI. Nat Biotechnol. 2023; 41:355–66. https://doi.org/10.1038/s41587-022-01487-9 [PubMed]
-
180.
Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015; 43:6557–67. https://doi.org/10.1093/nar/gkv596 [PubMed]
-
181.
Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–41. https://doi.org/10.1016/j.cell.2010.03.009 [PubMed]
-
182.
Yin HS, Zhou YL, Yang ZQ, Guo YL, Wang XX, Ai SY, Zhang XS. Electrochemical immunosensor for N6-methyladenosine RNA modification detection. Sensor Actuat B-Chem. 2015; 221:1–6. https://doi.org/10.1016/j.snb.2015.06.045
-
183.
Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021; 6:74. https://doi.org/10.1038/s41392-020-00450-x [PubMed]
-
184.
Tong J, Flavell RA, Li HB. RNA m6A modification and its function in diseases. Front Med. 2018; 12:481–9. https://doi.org/10.1007/s11684-018-0654-8 [PubMed]
-
185.
Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, Lan F, Shi YG, He C, et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018; 69:1028–38.e6. https://doi.org/10.1016/j.molcel.2018.02.015 [PubMed]
-
186.
Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m6A Transcripts by the 3'→5' RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol Cell. 2017; 68:374–87.e12. https://doi.org/10.1016/j.molcel.2017.09.021 [PubMed]
-
187.
Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017; 45:6051–63. https://doi.org/10.1093/nar/gkx141 [PubMed]
-
188.
Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015; 518:560–4. https://doi.org/10.1038/nature14234 [PubMed]
-
189.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, Schulman J, Famulare C, Patel M, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017; 23:1369–76. https://doi.org/10.1038/nm.4416 [PubMed]
-
190.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016; 62:335–45. https://doi.org/10.1016/j.molcel.2016.03.021 [PubMed]
-
191.
Xu H, Wang H, Zhao W, Fu S, Li Y, Ni W, Xin Y, Li W, Yang C, Bai Y, Zhan M, Lu L. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics. 2020; 10:5671–86. https://doi.org/10.7150/thno.42539 [PubMed]
-
192.
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y, Huang JT, Chen SM, Xu ZG, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018; 20:1074–83. https://doi.org/10.1038/s41556-018-0174-4 [PubMed]
-
193.
Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, Li X, Xu S, Miao J, Guo J, Zhang H, Gong J, Zhu F, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N6 adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020; 19:130. https://doi.org/10.1186/s12943-020-01249-8 [PubMed]
-
194.
Li F, Yi Y, Miao Y, Long W, Long T, Chen S, Cheng W, Zou C, Zheng Y, Wu X, Ding J, Zhu K, Chen D, et al. N6-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 2019; 79:5785–98. https://doi.org/10.1158/0008-5472.CAN-18-2868 [PubMed]
-
195.
Tassinari V, Cesarini V, Tomaselli S, Ianniello Z, Silvestris DA, Ginistrelli LC, Martini M, De Angelis B, De Luca G, Vitiani LR, Fatica A, Locatelli F, Gallo A. ADAR1 is a new target of METTL3 and plays a pro-oncogenic role in glioblastoma by an editing-independent mechanism. Genome Biol. 2021; 22:51. https://doi.org/10.1186/s13059-021-02271-9 [PubMed]
-
196.
Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, Duan P. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018; 151:356–65. https://doi.org/10.1016/j.ygyno.2018.09.015 [PubMed]
-
197.
Ying X, Jiang X, Zhang H, Liu B, Huang Y, Zhu X, Qi D, Yuan G, Luo J, Ji W. Programmable N6-methyladenosine modification of CDCP1 mRNA by RCas9-methyltransferase like 3 conjugates promotes bladder cancer development. Mol Cancer. 2020; 19:169. https://doi.org/10.1186/s12943-020-01289-0 [PubMed]
-
198.
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, Liu Y, Zhang X, Zhang W, Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018; 415:11–9. https://doi.org/10.1016/j.canlet.2017.11.018 [PubMed]
-
199.
Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016; 113:E2047–56. https://doi.org/10.1073/pnas.1602883113 [PubMed]
-
200.
Miao W, Chen J, Jia L, Ma J, Song D. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun. 2019; 516:719–25. https://doi.org/10.1016/j.bbrc.2019.06.128 [PubMed]
-
201.
Li J, Xie H, Ying Y, Chen H, Yan H, He L, Xu M, Xu X, Liang Z, Liu B, Wang X, Zheng X, Xie L. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer. 2020; 19:152. https://doi.org/10.1186/s12943-020-01267-6 [PubMed]
-
202.
Wang H, Wei W, Zhang ZY, Liu Y, Shi B, Zhong W, Zhang HS, Fang X, Sun CL, Wang JB, Liu LX. TCF4 and HuR mediated-METTL14 suppresses dissemination of colorectal cancer via N6-methyladenosine-dependent silencing of ARRDC4. Cell Death Dis. 2021; 13:3. https://doi.org/10.1038/s41419-021-04459-0 [PubMed]
-
203.
Wang K, Jiang L, Zhang Y, Chen C. Progression of Thyroid Carcinoma Is Promoted by the m6A Methyltransferase METTL3 Through Regulating m6A Methylation on TCF1. Onco Targets Ther. 2020; 13:1605–12. https://doi.org/10.2147/OTT.S234751 [PubMed]
-
204.
Xie JW, Huang XB, Chen QY, Ma YB, Zhao YJ, Liu LC, Wang JB, Lin JX, Lu J, Cao LL, Lin M, Tu RH, Zheng CH, et al. m6A modification-mediated BATF2 acts as a tumor suppressor in gastric cancer through inhibition of ERK signaling. Mol Cancer. 2020; 19:114. https://doi.org/10.1186/s12943-020-01223-4 [PubMed]
-
205.
Gong D, Zhang J, Chen Y, Xu Y, Ma J, Hu G, Huang Y, Zheng J, Zhai W, Xue W. The m6A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca2+ influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J Exp Clin Cancer Res. 2019; 38:233. https://doi.org/10.1186/s13046-019-1223-y [PubMed]
-
206.
Zhang P, He Q, Lei Y, Li Y, Wen X, Hong M, Zhang J, Ren X, Wang Y, Yang X, He Q, Ma J, Liu N. m6A-mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis. 2018; 9:1169. https://doi.org/10.1038/s41419-018-1224-3 [PubMed]
-
207.
Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y, Huang Y, Zheng R, Yu H, Wang J, Hu M, Miao J, Li J. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m6A-dependent manner. Cell Death Dis. 2020; 11:659. https://doi.org/10.1038/s41419-020-02847-6 [PubMed]
-
208.
Wang X, Tian L, Li Y, Wang J, Yan B, Yang L, Li Q, Zhao R, Liu M, Wang P, Sun Y. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. 2021; 40:80. https://doi.org/10.1186/s13046-021-01871-4 [PubMed]
-
209.
Zhuang C, Zhuang C, Luo X, Huang X, Yao L, Li J, Li Y, Xiong T, Ye J, Zhang F, Gui Y. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1α signalling axis. J Cell Mol Med. 2019; 23:2163–73. https://doi.org/10.1111/jcmm.14128 [PubMed]
-
210.
Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018; 57:590–7. https://doi.org/10.1002/mc.22782 [PubMed]
-
211.
Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018; 502:456–64. https://doi.org/10.1016/j.bbrc.2018.05.175 [PubMed]
-
212.
Li J, Zhu L, Shi Y, Liu J, Lin L, Chen X. m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am J Transl Res. 2019; 11:6084–92. [PubMed]
-
213.
Zheng X, Gong Y. Functions of RNA N6-methyladenosine modification in acute myeloid leukemia. Biomark Res. 2021; 9:36. https://doi.org/10.1186/s40364-021-00293-w [PubMed]
-
214.
Jiang Y, Wan Y, Gong M, Zhou S, Qiu J, Cheng W. RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-κB pathway. J Cell Mol Med. 2020; 24:6137–48. https://doi.org/10.1111/jcmm.15228 [PubMed]
-
215.
Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, Feng Y, Pan Q, Wan R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 2020; 19:91. https://doi.org/10.1186/s12943-020-01158-w [PubMed]
-
216.
Guo J, Wu Y, Du J, Yang L, Chen W, Gong K, Dai J, Miao S, Jin D, Xi S. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression. Oncogenesis. 2018; 7:49. https://doi.org/10.1038/s41389-018-0054-6 [PubMed]
-
217.
Zhu Z, Qian Q, Zhao X, Ma L, Chen P. N6-methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene. 2020; 731:144348. https://doi.org/10.1016/j.gene.2020.144348 [PubMed]
-
218.
Ma L, Chen T, Zhang X, Miao Y, Tian X, Yu K, Xu X, Niu Y, Guo S, Zhang C, Qiu S, Qiao Y, Fang W, et al. The m6A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2021; 38:101801. https://doi.org/10.1016/j.redox.2020.101801 [PubMed]
-
219.
Pi J, Wang W, Ji M, Wang X, Wei X, Jin J, Liu T, Qiang J, Qi Z, Li F, Liu Y, Ma Y, Si Y, et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res. 2021; 81:2651–65. https://doi.org/10.1158/0008-5472.CAN-20-0066 [PubMed]
-
220.
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, Xiao H, Li L, Rao S, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020; 48:3816–31. https://doi.org/10.1093/nar/gkaa048 [PubMed]
-
221.
Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJY, Xie Q, Vitting-Seerup K, Bhargava S, Dong Z, et al. The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 2021; 11:480–99. https://doi.org/10.1158/2159-8290.CD-20-0331 [PubMed]
-
222.
Chai RC, Chang YZ, Chang X, Pang B, An SY, Zhang KN, Chang YH, Jiang T, Wang YZ. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to activate NF-κB and promote the malignant progression of glioma. J Hematol Oncol. 2021; 14:109. https://doi.org/10.1186/s13045-021-01124-z [PubMed]
-
223.
Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, Zhou X, Wu X. FBW7 suppresses ovarian cancer development by targeting the N6-methyladenosine binding protein YTHDF2. Mol Cancer. 2021; 20:45. https://doi.org/10.1186/s12943-021-01340-8 [PubMed]
-
224.
Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K, Ji F, Ma Z, Hou B, He X. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene. 2020; 39:4507–18. https://doi.org/10.1038/s41388-020-1303-7 [PubMed]
-
225.
Müller S, Glaß M, Singh AK, Haase J, Bley N, Fuchs T, Lederer M, Dahl A, Huang H, Chen J, Posern G, Hüttelmaier S. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019; 47:375–90. https://doi.org/10.1093/nar/gky1012 [PubMed]
-
226.
Pu J, Wang J, Qin Z, Wang A, Zhang Y, Wu X, Wu Y, Li W, Xu Z, Lu Y, Tang Q, Wei H. IGF2BP2 Promotes Liver Cancer Growth Through an m6A-FEN1-Dependent Mechanism. Front Oncol. 2020; 10:578816. https://doi.org/10.3389/fonc.2020.578816 [PubMed]
-
227.
Xue T, Liu X, Zhang M, E Q, Liu S, Zou M, Li Y, Ma Z, Han Y, Thompson P, Zhang X. PADI2-Catalyzed MEK1 Citrullination Activates ERK1/2 and Promotes IGF2BP1-Mediated SOX2 mRNA Stability in Endometrial Cancer. Adv Sci (Weinh). 2021; 8:2002831. https://doi.org/10.1002/advs.202002831 [PubMed]
-
228.
Liu X, Su K, Sun X, Jiang Y, Wang L, Hu C, Zhang C, Lu M, Du X, Xing B. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/β-catenin pathway. J Exp Clin Cancer Res. 2021; 40:132. https://doi.org/10.1186/s13046-021-01934-6 [PubMed]
-
229.
Regué L, Zhao L, Ji F, Wang H, Avruch J, Dai N. RNA m6A reader IMP2/IGF2BP2 promotes pancreatic β-cell proliferation and insulin secretion by enhancing PDX1 expression. Mol Metab. 2021; 48:101209. https://doi.org/10.1016/j.molmet.2021.101209 [PubMed]
-
230.
Yang Z, Wang T, Wu D, Min Z, Tan J, Yu B. RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J Exp Clin Cancer Res. 2020; 39:203. https://doi.org/10.1186/s13046-020-01714-8 [PubMed]
-
231.
Dahal U, Le K, Gupta M. RNA m6A methyltransferase METTL3 regulates invasiveness of melanoma cells by matrix metallopeptidase 2. Melanoma Res. 2019; 29:382–9. https://doi.org/10.1097/CMR.0000000000000580 [PubMed]
-
232.
Jiang F, Tang X, Tang C, Hua Z, Ke M, Wang C, Zhao J, Gao S, Jurczyszyn A, Janz S, Beksac M, Zhan F, Gu C, Yang Y. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J Hematol Oncol. 2021; 14:54. https://doi.org/10.1186/s13045-021-01066-6 [PubMed]
-
233.
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, Liang X, Yang Y. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021; 28:335–49. https://doi.org/10.1038/s41417-020-00222-3 [PubMed]
-
234.
Pan X, Hong X, Li S, Meng P, Xiao F. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med. 2021; 53:91–102. https://doi.org/10.1038/s12276-020-00510-w [PubMed]
-
235.
He H, Wu W, Sun Z, Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of SEC62. Biochem Biophys Res Commun. 2019; 517:581–7. https://doi.org/10.1016/j.bbrc.2019.07.058 [PubMed]
-
236.
Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, Wang D, Huang J, Gao S, Gao Y. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017; 482:582–9. https://doi.org/10.1016/j.bbrc.2016.11.077 [PubMed]
-
237.
Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019; 11:1177–87. https://doi.org/10.2147/CMAR.S181058 [PubMed]
-
238.
Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, Zhao W, Fan Y, Zhang D, Sun R. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020; 53:e12768. https://doi.org/10.1111/cpr.12768 [PubMed]
-
239.
Yue C, Chen J, Li Z, Li L, Chen J, Guo Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J Exp Clin Cancer Res. 2020; 39:240. https://doi.org/10.1186/s13046-020-01731-7 [PubMed]
-
240.
Shen XP, Ling X, Lu H, Zhou CX, Zhang JK, Yu Q. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur Rev Med Pharmacol Sci. 2018; 22:8220–6. https://doi.org/10.26355/eurrev_201812_16516 [PubMed]
-
241.
Kleemann M, Schneider H, Unger K, Sander P, Schneider EM, Fischer-Posovszky P, Handrick R, Otte K. MiR-744-5p inducing cell death by directly targeting HNRNPC and NFIX in ovarian cancer cells. Sci Rep. 2018; 8:9020. https://doi.org/10.1038/s41598-018-27438-6 [PubMed]
-
242.
Xu C, Yuan B, He T, Ding B, Li S. Prognostic values of YTHDF1 regulated negatively by mir-3436 in Glioma. J Cell Mol Med. 2020; 24:7538–49. https://doi.org/10.1111/jcmm.15382 [PubMed]
-
243.
Yao FY, Zhao C, Zhong FM, Qin TY, Wen F, Li MY, Liu J, Huang B, Wang XZ. m6A Modification of lncRNA NEAT1 Regulates Chronic Myelocytic Leukemia Progression via miR-766-5p/CDKN1A Axis. Front Oncol. 2021; 11:679634. https://doi.org/10.3389/fonc.2021.679634 [PubMed]
-
244.
Rong D, Dong Q, Qu H, Deng X, Gao F, Li Q, Sun P. m6A-induced LINC00958 promotes breast cancer tumorigenesis via the miR-378a-3p/YY1 axis. Cell Death Discov. 2021; 7:27. https://doi.org/10.1038/s41420-020-00382-z [PubMed]
-
245.
Liu T, Wang H, Fu Z, Wang Z, Wang J, Gan X, Wang A, Wang L. Methyltransferase-like 14 suppresses growth and metastasis of renal cell carcinoma by decreasing long noncoding RNA NEAT1. Cancer Sci. 2022; 113:446–58. https://doi.org/10.1111/cas.15212 [PubMed]
-
246.
Guo T, Liu DF, Peng SH, Xu AM. ALKBH5 promotes colon cancer progression by decreasing methylation of the lncRNA NEAT1. Am J Transl Res. 2020; 12:4542–9. [PubMed]
-
247.
Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y, Mei Y, Tan Y, Li X, Zeng Z, Xiong W, Li G, Li X, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020; 14:1282–96. https://doi.org/10.1002/1878-0261.12676 [PubMed]
-
248.
Ji F, Lu Y, Chen S, Lin X, Yu Y, Zhu Y, Luo X. m6A methyltransferase METTL3-mediated lncRNA FOXD2-AS1 promotes the tumorigenesis of cervical cancer. Mol Ther Oncolytics. 2021; 22:574–81. https://doi.org/10.1016/j.omto.2021.07.004 [PubMed]
-
249.
Liu HT, Zou YX, Zhu WJ, Sen-Liu, Zhang GH, Ma RR, Guo XY, Gao P. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. 2022; 29:627–41. https://doi.org/10.1038/s41418-021-00879-9 [PubMed]
-
250.
Wu J, Pang R, Li M, Chen B, Huang J, Zhu Y. m6A-Induced LncRNA MEG3 Suppresses the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cell Through miR-544b/BTG2 Signaling. Onco Targets Ther. 2021; 14:3745–55. https://doi.org/10.2147/OTT.S289198 [PubMed]
-
251.
Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z, Du H, Ren D, Dai Y, Peng X. m6 A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med. 2021; 11:e426. https://doi.org/10.1002/ctm2.426 [PubMed]
-
252.
Qian X, Yang J, Qiu Q, Li X, Jiang C, Li J, Dong L, Ying K, Lu B, Chen E, Liu P, Lu Y. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J Hematol Oncol. 2021; 14:112. https://doi.org/10.1186/s13045-021-01123-0 [PubMed]
-
253.
Shen D, Ding L, Lu Z, Wang R, Yu C, Wang H, Zheng Q, Wang X, Xu W, Yu H, Xu L, Wang M, Yu S, et al. METTL14-mediated Lnc-LSG1 m6A modification inhibits clear cell renal cell carcinoma metastasis via regulating ESRP2 ubiquitination. Mol Ther Nucleic Acids. 2021; 27:547–61. https://doi.org/10.1016/j.omtn.2021.12.024 [PubMed]
-
254.
Zhang X, Li D, Jia C, Cai H, Lv Z, Wu B. METTL14 promotes tumorigenesis by regulating lncRNA OIP5-AS1/miR-98/ADAMTS8 signaling in papillary thyroid cancer. Cell Death Dis. 2021; 12:617. https://doi.org/10.1038/s41419-021-03891-6 [PubMed]
-
255.
Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, Lu S, Xu D, Wu Y, Chen Q, Ding X, Guo H, Li Y, et al. LNC942 promoting METTL14-mediated m6A methylation in breast cancer cell proliferation and progression. Oncogene. 2020; 39:5358–72. https://doi.org/10.1038/s41388-020-1338-9 [PubMed]
-
256.
Wang J, Zha J, Wang X. Knockdown of lncRNA NUTM2A-AS1 inhibits lung adenocarcinoma cell viability by regulating the miR-590-5p/METTL3 axis. Oncol Lett. 2021; 22:798. https://doi.org/10.3892/ol.2021.13059 [PubMed]
-
257.
Wang G, Zhang Z, Xia C. Long non-coding RNA LINC00240 promotes gastric cancer progression via modulating miR-338-5p/METTL3 axis. Bioengineered. 2021; 12:9678–91. https://doi.org/10.1080/21655979.2021.1983276 [PubMed]
-
258.
Hu H, Kong Q, Huang XX, Zhang HR, Hu KF, Jing Y, Jiang YF, Peng Y, Wu LC, Fu QS, Xu L, Xia YB. Longnon-coding RNA BLACAT2 promotes gastric cancer progression via the miR-193b-5p/METTL3 pathway. J Cancer. 2021; 12:3209–21. https://doi.org/10.7150/jca.50403 [PubMed]
-
259.
Bo C, Li N, He L, Zhang S, An Y. Long non-coding RNA ILF3-AS1 facilitates hepatocellular carcinoma progression by stabilizing ILF3 mRNA in an m6A-dependent manner. Hum Cell. 2021; 34:1843–54. https://doi.org/10.1007/s13577-021-00608-x [PubMed]
-
260.
Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019; 11:4909–21. [PubMed]
-
261.
Tang S, Liu Q, Xu M. LINC00857 promotes cell proliferation and migration in colorectal cancer by interacting with YTHDC1 and stabilizing SLC7A5. Oncol Lett. 2021; 22:578. https://doi.org/10.3892/ol.2021.12839 [PubMed]
-
262.
Wang C, Gu Y, Zhang E, Zhang K, Qin N, Dai J, Zhu M, Liu J, Xie K, Jiang Y, Guo X, Liu M, Jin G, et al. A cancer-testis non-coding RNA LIN28B-AS1 activates driver gene LIN28B by interacting with IGF2BP1 in lung adenocarcinoma. Oncogene. 2019; 38:1611–24. https://doi.org/10.1038/s41388-018-0548-x [PubMed]
-
263.
Wang H, Liang L, Dong Q, Huan L, He J, Li B, Yang C, Jin H, Wei L, Yu C, Zhao F, Li J, Yao M, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018; 8:2814–29. https://doi.org/10.7150/thno.23012 [PubMed]
-
264.
Gu Y, Niu S, Wang Y, Duan L, Pan Y, Tong Z, Zhang X, Yang Z, Peng B, Wang X, Han X, Li Y, Cheng T, et al. DMDRMR-Mediated Regulation of m6A-Modified CDK4 by m6A Reader IGF2BP3 Drives ccRCC Progression. Cancer Res. 2021; 81:923–34. https://doi.org/10.1158/0008-5472.CAN-20-1619 [PubMed]
-
265.
Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang J, Lu B, Yuan L, Sun M, Wang Z. Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol Ther. 2020; 28:1479–93. https://doi.org/10.1016/j.ymthe.2020.03.010 [PubMed]
-
266.
Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, Gorshkov K, Mao Q, Xia S, Cen D, Zheng J, Liang X, Cai X. N6-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. 2020; 19:163. https://doi.org/10.1186/s12943-020-01281-8 [PubMed]
-
267.
Wang T, Liu Z, She Y, Deng J, Zhong Y, Zhao M, Li S, Xie D, Sun X, Hu X, Chen C. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Lett. 2021; 520:321–31. https://doi.org/10.1016/j.canlet.2021.08.007 [PubMed]
-
268.
Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X, Fan Y. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021; 12:298. https://doi.org/10.1038/s41419-021-03558-2 [PubMed]
-
269.
Wang M, Yang Y, Yang J, Yang J, Han S. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m6A-YTHDF3-Zeb1. Life Sci. 2020; 257:118082. https://doi.org/10.1016/j.lfs.2020.118082 [PubMed]
-
270.
Jiang X, Xing L, Chen Y, Qin R, Song S, Lu Y, Xie S, Wang L, Pu H, Gui X, Li T, Xu J, Li J, et al. CircMEG3 inhibits telomerase activity by reducing Cbf5 in human liver cancer stem cells. Mol Ther Nucleic Acids. 2020; 23:310–23. https://doi.org/10.1016/j.omtn.2020.11.009 [PubMed]
-
271.
Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, Ma X, Cheng Z, Yu C, Wang S, Zuo Q, Zhou Y, Wang J, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021; 12:295. https://doi.org/10.1038/s41467-020-20527-z [PubMed]
-
272.
Xie F, Huang C, Liu F, Zhang H, Xiao X, Sun J, Zhang X, Jiang G. CircPTPRA blocks the recognition of RNA N6-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer. 2021; 20:68. https://doi.org/10.1186/s12943-021-01359-x [PubMed]
-
273.
Zheng Y, Nie P, Peng D, He Z, Liu M, Xie Y, Miao Y, Zuo Z, Ren J. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res. 2018; 46:D139–45. https://doi.org/10.1093/nar/gkx895 [PubMed]
-
274.
Han Y, Feng J, Xia L, Dong X, Zhang X, Zhang S, Miao Y, Xu Q, Xiao S, Zuo Z, Xia L, He C. CVm6A: A Visualization and Exploration Database for m6As in Cell Lines. Cells. 2019; 8:168. https://doi.org/10.3390/cells8020168 [PubMed]
-
275.
Wu X, Wei Z, Chen K, Zhang Q, Su J, Liu H, Zhang L, Meng J. m6Acomet: large-scale functional prediction of individual m6A RNA methylation sites from an RNA co-methylation network. BMC Bioinformatics. 2019; 20:223. https://doi.org/10.1186/s12859-019-2840-3 [PubMed]
-
276.
Zhou D, Wang H, Bi F, Xing J, Gu Y, Wang C, Zhang M, Huang Y, Zeng J, Wu Q, Zhang Y. M6ADD: a comprehensive database of m6A modifications in diseases. RNA Biol. 2021; 18:2354–62. https://doi.org/10.1080/15476286.2021.1913302 [PubMed]
-
277.
Liu Q, Gregory RI. RNAmod: an integrated system for the annotation of mRNA modifications. Nucleic Acids Res. 2019; 47:W548–55. https://doi.org/10.1093/nar/gkz479 [PubMed]
-
278.
Tang Y, Chen K, Wu X, Wei Z, Zhang SY, Song B, Zhang SW, Huang Y, Meng J. DRUM: Inference of Disease-Associated m6A RNA Methylation Sites From a Multi-Layer Heterogeneous Network. Front Genet. 2019; 10:266. https://doi.org/10.3389/fgene.2019.00266 [PubMed]
-
279.
Nie F, Feng P, Song X, Wu M, Tang Q, Chen W. RNAWRE: a resource of writers, readers and erasers of RNA modifications. Database (Oxford). 2020; 2020:baaa049. https://doi.org/10.1093/database/baaa049 [PubMed]
-
280.
Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016; 44:e91. https://doi.org/10.1093/nar/gkw104 [PubMed]
-
281.
Antanaviciute A, Baquero-Perez B, Watson CM, Harrison SM, Lascelles C, Crinnion L, Markham AF, Bonthron DT, Whitehouse A, Carr IM. m6aViewer: software for the detection, analysis, and visualization of N6-methyladenosine peaks from m6A-seq/ME-RIP sequencing data. RNA. 2017; 23:1493–501. https://doi.org/10.1261/rna.058206.116 [PubMed]
-
282.
Xiang S, Liu K, Yan Z, Zhang Y, Sun Z. RNAMethPre: A Web Server for the Prediction and Query of mRNA m6A Sites. PLoS One. 2016; 11:e0162707. https://doi.org/10.1371/journal.pone.0162707 [PubMed]
-
283.
Chen K, Wei Z, Zhang Q, Wu X, Rong R, Lu Z, Su J, de Magalhães JP, Rigden DJ, Meng J. WHISTLE: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acids Res. 2019; 47:e41. https://doi.org/10.1093/nar/gkz074 [PubMed]
-
284.
Xiang S, Yan Z, Liu K, Zhang Y, Sun Z. AthMethPre: a web server for the prediction and query of mRNA m6A sites in Arabidopsis thaliana. Mol Biosyst. 2016; 12:3333–7. https://doi.org/10.1039/c6mb00536e [PubMed]
-
285.
Wang X, Yan R. RFAthM6A: a new tool for predicting m6A sites in Arabidopsis thaliana. Plant Mol Biol. 2018; 96:327–37. https://doi.org/10.1007/s11103-018-0698-9 [PubMed]
-
286.
Wang M, Xie J, Xu S. M6A-BiNP: predicting N6-methyladenosine sites based on bidirectional position-specific propensities of polynucleotides and pointwise joint mutual information. RNA Biol. 2021; 18:2498–512. https://doi.org/10.1080/15476286.2021.1930729 [PubMed]
-
287.
Qiang X, Chen H, Ye X, Su R, Wei L. M6AMRFS: Robust Prediction of N6-Methyladenosine Sites With Sequence-Based Features in Multiple Species. Front Genet. 2018; 9:495. https://doi.org/10.3389/fgene.2018.00495 [PubMed]
-
288.
Zhang L, Qin X, Liu M, Xu Z, Liu G. DNN-m6A: A Cross-Species Method for Identifying RNA N6-Methyladenosine Sites Based on Deep Neural Network with Multi-Information Fusion. Genes (Basel). 2021; 12:354. https://doi.org/10.3390/genes12030354 [PubMed]
-
289.
Liang Z, Zhang L, Chen H, Huang D, Song B. m6A-Maize: Weakly supervised prediction of m6A-carrying transcripts and m6A-affecting mutations in maize (Zea mays). Methods. 2022; 203:226–32. https://doi.org/10.1016/j.ymeth.2021.11.010 [PubMed]
-
290.
Abbas Z, Tayara H, Zou Q, Chong KT. TS-m6A-DL: Tissue-specific identification of N6-methyladenosine sites using a universal deep learning model. Comput Struct Biotechnol J. 2021; 19:4619–25. https://doi.org/10.1016/j.csbj.2021.08.014 [PubMed]
-
291.
Dao FY, Lv H, Yang YH, Zulfiqar H, Gao H, Lin H. Computational identification of N6-methyladenosine sites in multiple tissues of mammals. Comput Struct Biotechnol J. 2020; 18:1084–91. https://doi.org/10.1016/j.csbj.2020.04.015 [PubMed]
-
292.
Wang Y, Guo R, Huang L, Yang S, Hu X, He K. m6AGE: A Predictor for N6-Methyladenosine Sites Identification Utilizing Sequence Characteristics and Graph Embedding-Based Geometrical Information. Front Genet. 2021; 12:670852. https://doi.org/10.3389/fgene.2021.670852 [PubMed]
-
293.
Zhang S, Wang J, Li X, Liang Y. M6A-GSMS: Computational identification of N6-methyladenosine sites with GBDT and stacking learning in multiple species. J Biomol Struct Dyn. 2022; 40:12380–91. https://doi.org/10.1080/07391102.2021.1970628 [PubMed]
-
294.
Chen W, Feng P, Yang H, Ding H, Lin H, Chou KC. iRNA-3typeA: Identifying Three Types of Modification at RNA's Adenosine Sites. Mol Ther Nucleic Acids. 2018; 11:468–74. https://doi.org/10.1016/j.omtn.2018.03.012 [PubMed]
-
295.
Chen W, Feng P, Ding H, Lin H, Chou KC. iRNA-Methyl: Identifying N(6)-methyladenosine sites using pseudo nucleotide composition. Anal Biochem. 2015; 490:26–33. https://doi.org/10.1016/j.ab.2015.08.021 [PubMed]
-
296.
Huang Y, He N, Chen Y, Chen Z, Li L. BERMP: a cross-species classifier for predicting m6A sites by integrating a deep learning algorithm and a random forest approach. Int J Biol Sci. 2018; 14:1669–77. https://doi.org/10.7150/ijbs.27819 [PubMed]
-
297.
Wei L, Chen H, Su R. M6APred-EL: A Sequence-Based Predictor for Identifying N6-methyladenosine Sites Using Ensemble Learning. Mol Ther Nucleic Acids. 2018; 12:635–44. https://doi.org/10.1016/j.omtn.2018.07.004 [PubMed]
-
298.
Chen W, Feng P, Ding H, Lin H. Identifying N 6-methyladenosine sites in the Arabidopsis thaliana transcriptome. Mol Genet Genomics. 2016; 291:2225–9. https://doi.org/10.1007/s00438-016-1243-7 [PubMed]
-
299.
Li GQ, Liu Z, Shen HB, Yu DJ. TargetM6A: Identifying N6-Methyladenosine Sites From RNA Sequences via Position-Specific Nucleotide Propensities and a Support Vector Machine. IEEE Trans Nanobioscience. 2016; 15:674–82. https://doi.org/10.1109/TNB.2016.2599115 [PubMed]
-
300.
Liu Z, Xiao X, Yu DJ, Jia J, Qiu WR, Chou KC. pRNAm-PC: Predicting N(6)-methyladenosine sites in RNA sequences via physical-chemical properties. Anal Biochem. 2016; 497:60–7. https://doi.org/10.1016/j.ab.2015.12.017 [PubMed]
-
301.
Zhang M, Sun JW, Liu Z, Ren MW, Shen HB, Yu DJ. Improving N(6)-methyladenosine site prediction with heuristic selection of nucleotide physical-chemical properties. Anal Biochem. 2016; 508:104–13. https://doi.org/10.1016/j.ab.2016.06.001 [PubMed]
-
302.
Chen W, Xing P, Zou Q. Detecting N6-methyladenosine sites from RNA transcriptomes using ensemble Support Vector Machines. Sci Rep. 2017; 7:40242. https://doi.org/10.1038/srep40242 [PubMed]
-
303.
Feng P, Ding H, Yang H, Chen W, Lin H, Chou KC. iRNA-PseColl: Identifying the Occurrence Sites of Different RNA Modifications by Incorporating Collective Effects of Nucleotides into PseKNC. Mol Ther Nucleic Acids. 2017; 7:155–63. https://doi.org/10.1016/j.omtn.2017.03.006 [PubMed]
-
304.
Xing P, Su R, Guo F, Wei L. Identifying N6-methyladenosine sites using multi-interval nucleotide pair position specificity and support vector machine. Sci Rep. 2017; 7:46757. https://doi.org/10.1038/srep46757 [PubMed]
-
305.
Akbar S, Hayat M. iMethyl-STTNC: Identification of N6-methyladenosine sites by extending the idea of SAAC into Chou's PseAAC to formulate RNA sequences. J Theor Biol. 2018; 455:205–11. https://doi.org/10.1016/j.jtbi.2018.07.018 [PubMed]
-
306.
Zou Q, Xing P, Wei L, Liu B. Gene2vec: gene subsequence embedding for prediction of mammalian N6-methyladenosine sites from mRNA. RNA. 2019; 25:205–18. https://doi.org/10.1261/rna.069112.118 [PubMed]
-
307.
Chen W, Ding H, Zhou X, Lin H, Chou KC. iRNA(m6A)-PseDNC: Identifying N6-methyladenosine sites using pseudo dinucleotide composition. Anal Biochem. 2018; 561–562:59–65. https://doi.org/10.1016/j.ab.2018.09.002 [PubMed]
-
308.
Song B, Chen K, Tang Y, Wei Z, Su J, de Magalhães JP, Rigden DJ, Meng J. ConsRM: collection and large-scale prediction of the evolutionarily conserved RNA methylation sites, with implications for the functional epitranscriptome. Brief Bioinform. 2021; 22:bbab088. https://doi.org/10.1093/bib/bbab088 [PubMed]
-
309.
Zhang SY, Zhang SW, Tang Y, Fan XN, Meng J. Funm6AViewer: a web server and R package for functional analysis of context-specific m6A RNA methylation. Bioinformatics. 2021; 37:4277–9. https://doi.org/10.1093/bioinformatics/btab362 [PubMed]
-
310.
Pratanwanich PN, Yao F, Chen Y, Koh CWQ, Wan YK, Hendra C, Poon P, Goh YT, Yap PML, Chooi JY, Chng WJ, Ng SB, Thiery A, et al. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat Biotechnol. 2021; 39:1394–402. https://doi.org/10.1038/s41587-021-00949-w [PubMed]
-
311.
Xiong Y, He X, Zhao D, Tian T, Hong L, Jiang T, Zeng J. Modeling multi-species RNA modification through multi-task curriculum learning. Nucleic Acids Res. 2021; 49:3719–34. https://doi.org/10.1093/nar/gkab124 [PubMed]
-
312.
Leger A, Amaral PP, Pandolfini L, Capitanchik C, Capraro F, Miano V, Migliori V, Toolan-Kerr P, Sideri T, Enright AJ, Tzelepis K, van Werven FJ, Luscombe NM, et al. RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat Commun. 2021; 12:7198. https://doi.org/10.1038/s41467-021-27393-3 [PubMed]
-
313.
Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat Protoc. 2014; 9:828–41. https://doi.org/10.1038/nprot.2014.047 [PubMed]
-
314.
Yuen KC, Xu B, Krantz ID, Gerton JL. NIPBL Controls RNA Biogenesis to Prevent Activation of the Stress Kinase PKR. Cell Rep. 2016; 14:93–102. https://doi.org/10.1016/j.celrep.2015.12.012 [PubMed]