Age-associated epigenetic modifications in human DNA increase its immunogenicity
Abstract
Chronic inflammation, increased reactivity to self-antigens and incidences of cancer are hallmarks of aging. However, the underlying mechanisms are not well understood. Age-associated alterations in the DNA either due to oxidative damage, defects in DNA repair or epigenetic modifications such as methylation that lead to mutations and changes in the expression of genes are thought to be partially responsible. Here we report that epigenetic modifications in aged DNA also increase its immunogenicity rendering it more reactive to innate immune system cells such as the dendritic cells. We observed increased upregulation of costimulatory molecules as well as enhanced secretion of IFN-α from dendritic cells in response to DNA from aged donors as compared to DNA from young donors when it was delivered intracellularly via Lipofectamine. Investigations into the mechanisms revealed that DNA from aged subjects is not degraded, neither is it more damaged compared to DNA from young subjects. However, there is significantly decreased global level of methylation suggesting that age-associated hypomethylation of the DNA may be the cause of its increased immunogenicity. Increased immunogenicity of self DNA may thus be another mechanism that may contribute to the increase in age-associated chronic inflammation, autoimmunity and cancer.
Introduction
Chronic inflammation and increased self
reactivity are the hallmarks of aging and are thought to be the underlying
cause of the many age-associated diseases such as Alzheimer's; Rheumatoid
Arthritis [1]. The mechanisms responsible are not well understood. It is well
established that the immune system undergoes significant alterations with age.
The consequences of progressive aging of the immune system include an increase
in autoimmune phenomena, incidence of cancer, chronic inflammation, and
predisposition to infections [2-5]. This suggests a decrease in the protective
immune responses to exogenous and infectious agents, and an increase in
reactivity to self or endogenous danger signals. Several factors may be responsible
for the increased reactivity to self, including loss of immune tolerance, and
progressive age-associated loss of tissue integrity yielding new self-antigens.
Another possibility is that advancing age leads to modifications in existing
self-antigens [3,4,6] rendering them more immunogenic.
Human
DNA is an example of self-antigen that undergoes age-associated genetic and
epigenetic alterations [7-11]. It is well documented that aging cells subtly
change their patterns of DNA methylation. Overall, cells and tissues become
hypomethylated while selected genes become progressively hypermethylated and,
potentially, permanently silenced [12,13]. This is believed to be poor
prognosis for malignancies [14,15]. A large body of experimental evidence also
supports the existence of a relationship between genomic instability, DNA
damage and aging [10]. DNA repair functions are severely compromised with age
leading to DNA lesions with single and double stranded breaks [15,16].
Moreover, free radicals and reactive oxygen species cause further damage to the
DNA and oxidative DNA damage has been implicated in carcinogenesis, ageing
[10,11,17] and several age-related degenerative diseases [18]. More recently
Rodier et al. [19] have shown that persistent DNA damage in senescent cells
induces pro-inflammatory cytokine secretion. It is thus clear that aging leads
to alterations in DNA that are implicated in various age-related disorders.
In
the present study, we have determined if the age-associated alterations in DNA
also affect its immunogenicity. We have tested our hypothesis using a
previously established system [20] whereby we deliver human DNA to dendritic
cells using transfection agent, Lipofectamine.
Results
DNA
from aged subjects is more immunogenic than DNA from young subjects
Human DNA is
generally inert and does not stimulate dendritic cells (DCs). However, recent
studies from our laboratory [20] as well as from others [21-23] suggest that
DNA can activate DCs if delivered in vitro inside the cell through
either transfection or via immune complexes. In vitro delivery of DNA
mimics the normal physiological conditions where human DNA gains entry into the
cells as complexed with proteins or cell debris from dying cells or immune
complexes. Therefore, to investigate our hypothesis, DNA extracted from the
blood of aged and young subjects was delivered inside human monocyte derived
DCs from healthy young donors using the transfection reagent lipofectamine. The
concentration of DNA used was 1 μg/ml. This was found to be the optimal concentration from
our previous studies [20] since higher concentrations were toxic while lower
concentrations led to insufficient activation of DCs. Delivery of DNA led to
the activation of DCs as evidenced by upregulation of costimulatory markers
CD80 and CD86 (Figure 1A and B) and secretion of cytokines IFN-α (Figure 1C). A significantly
increased upregulation of CD80 (p=0.008) and CD86 (p=0.02) was observed in
aged DNA (data is of 15 separate aged and young DNA) stimulated DCs compared to
DCs stimulated with young DNA (Figure 1A and B). The maturation marker CD83 was
not significantly upregulated (p=0.29) in either group. Cytokine
profile shows that there was significantly increased (p=0.003) secretion of
IFN-α (data is of
30 separate aged and young DNA, Figure1C) by aged DNA-treated DCs as compared
to young DNA-treated DCs. Introduction of
lipofectamine alone did not activate DCs.Stimulatory
activity of the DNA was lost when delivered without lipofectamine suggesting
that exposure to DNA alone does not induce activation of DCs and that this
requires its intracellular delivery. These data suggest that aging leads
to alterations in DNA that enhance its immunogenicity, and may contribute to
age-associated chronic inflammation and autoimmune phenomenon.
DNA
from aged subjects is demethylated compared to DNA from young subjects
Next,
we investigated the age-associated alterations in the DNA that may be
responsible for its increased immunogenicity. Aging is associated with
increased apoptosis [24], which may result in shorter fragments of DNA. Running
the DNA on gel showed that the size of DNA obtained from both aged and young
subjects was comparable (Figure 2A). Accumulation of DNA damage due to either
oxidation or inefficient DNA repair mechanism is another characteristic of
advancing age [15,16,25]. Therefore, we compared the DNA damage between aged and
young DNA [damage of 25 separate aged and young DNA was determined] by
measuring the number of abasic sites by ELISA. Indeed, we observed an increase
in DNA damage in the DNA from aged subjects relative to DNA from young;
however, the difference was not significant (p=0.27, Figure 2B). Age-associated
changes in DNA methylation patterns are also a hallmark of aging [8-10,26-29];
both an increase and a decrease DNA methylation have been reported [10,29]. To
determine if modifications in methylation are responsible for the
immunogenicity of aged DNA, we compared the methylation level ofthe
DNA from aged and young subjects using the global DNA methylation ELISA kit
(methylation levels of 24 separate aged and young DNA were determined). In
this kit the methylated fraction of DNA is recognized by 5-methylcytosine
antibody and quantified through an ELISA-like reaction. There was an
approximately ten percent decrease (p=0.001) of DNA methylation in aged as
compared to the DNA from young (Figure 2C). This suggests that DNA from aged is
hypomethylated, which may result in enhancing its immunogenicity.
Immunogenicity
of mammalian DNA correlates inversely with DNA methylation
To
further confirm if hypomethylation of aged DNA influencesthe
immunogenicity of the DNA, we in vitro methylated the DNA from aged
subjects using a methyl transferase
enzyme. The percent of DNA methylation correlated with time with increased
methylation being observed at 18h compared to 4h of the reaction (Figure 3A).The experiment was repeated with five separate DNAs. In two experiments
commercially obtained DNA from Jurkat and Hela cell lines were used for
methylation. This was done to confirm that the results obtained are not an
artifact of our purification process.
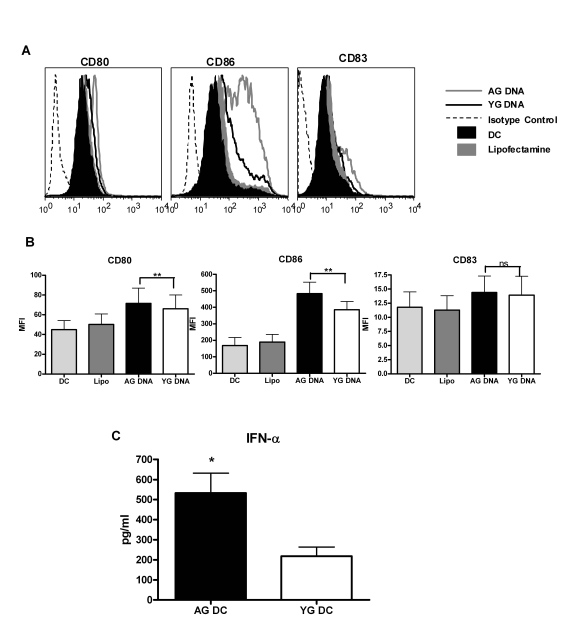
Figure 1. DNA from aged subjects is more immunogenic than DNA from young subjects. (A) DCs
were activated with aged and young DNA complexed with lipofectamine. The
expression of costimulatory molecules CD80 and CD86 and the maturation
molecule (CD83) in the unactivated and activated DCs was measured by flow
cytometry. Figure is representative of ten such experiments using fifteen
separate aged and young DNA. (B) Bar graph represents the
mean fluorescence intensity of CD80, CD86 and CD83 of the same. (C)
Supernatants collected from DCs activated with aged and young DNA were
assayed for IFN-α using specific
ELISA. Bar diagrams depict the concentration of IFN-α secreted by the DCs. Figure is
mean +
S.E. of thirty separate aged and young DNA tested.
Furthermore,
intracellular delivery of this methylated DNA into DCs resulted in decrease in
IFN-α secretion compared to untreated DNA control (Figure 3B). The decrease in IFN-α production correlated with increase in DNA
methylation. These data clearly demonstrate that epigenetic changes in the DNA
from aged subjects' leads to decreased DNA methylation resulting in an enhanced
immunogenicity of the DNA.
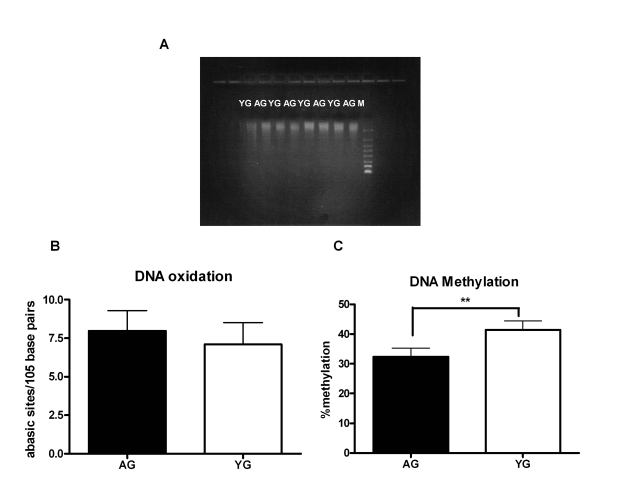
Figure 2. DNA from aged subjects is demethylated compared to DNA from young subjects. (A) FlashGel
showing the molecular weight of Aged and young DNA. Figure is
representative of eight such experiments. (B) Bar diagram
depicting the damage in DNA from aged and young subjects as determined by
ELISA that measures the number of abasic sites per 105 base
pairs. Figure is mean +
S.E. of twenty five separate aged and young
DNA tested. (C) Bar diagram depicts the percent of global
methylation in aged and young DNA as measured by ELISA. The methylated
fraction of DNA is recognized by 5-methylcytosine antibody and quantified
through an ELISA-like reaction. Figure is mean +
S.E. of twenty four
separate aged and young DNA tested.
Since
our results demonstrated a small but insignificant increased damage in the DNA
from aged, we investigated if this damage also affected the immunogenicity of
the DNA. PBMCs from young subjects were exposed to hyderogen peroxide to
induce DNA damage. This treatment led to an increased DNA damage as shown in
Figure 3C; however, intracellular delivery of this damaged DNA into DCs did not
result in increased activation compared to undamaged DNA from the same
individual (Figure 3D). This suggests that DNA damage does not alter its
immunogenicity further confirming that hypomethylation of the DNA is
responsible for its increased immunogenicity.
Discussion
Decreases in global level of methylation
along with a concomitant increase in promoter methylation are the hallmark of
age-associated epigenetic changes [10,29].
These
age-associated epigenetic changes are thought to play a key role in the
development of cancer and autoimmune phenomena through modification of gene
expression. Causes for age-related methylation changes remain unknown.
Accumulating
studies have indicated a potential role of DNAhypomethylation in
the pathogenesis of autoimmune diseases [30-36]. Earlier in vivo studies
have shown potentiation of autoimmunity in mice treated with hypomethylating
agents such as5-azacytidine, procainamide and hydralazine [31,34].Others studies
described that DNA hypomethylation
is essential for apoptotic DNA to induceSLE-like autoimmune disease
in non-susceptible mice [35]. Changes in human DNA methylation patterns are
also an important feature of cancer development and progression [14,33,36,37]. Alterations
of DNA
methylation are one of the most consistentepigenetic
changes in
human cancers [37]. Human cancers generallyshow global DNA
hypomethylation
accompanied by region-specifichypermethylation a pattern similar to
aging. Studies have shown hypomethylation of squamous cell carcinomas in White
men was associated with shorter survival from the disease [38]. A potential role of DNA
hypomethylation in other conditions such as atherosclerosis and autoimmune
diseases [e.g., multiple sclerosis and lupus] is also being recognized [39,40].
As is evident abnormal DNA methylation plays a very
important role in various pathologic states, such as leukemia and autoimmunity.
The underlying mechanisms are however not fully delineated so far. Our data
suggests that along with regulating the transcription of various genes these
methylation changes also render the DNA to be more immunogenic. The immune system is normally protected from exposure to
self dsDNA during apoptosis due to the rapid engulfment of apoptotic cells, and the abundance
of extra- and intracellular DNases [41,42]. However, phagocytic cells may be exposed to cellular DNA
following tissue necrosis, inflammation, or viral infection. Defective
clearance of apoptotic cells would also result in an accumulation of late phase
apoptotic cells. Previous study from our laboratory in humans [43] and a recent study [44] in mice suggest that apoptotic cell clearance is decreased with
age. This may result in release of DNA from apoptotic cells due to secondary
necrosis. Such DNA in aging would be much more immunogenic since it is
hypomethylated and would lead to maturation of dendritic cells and increased
reactivity to self resulting in slow loss of peripheral self tolerance. The increased immunogenicity of hypomethylated DNA may
thus be one of the mechanisms that contribute to the development of
autoreactivity, cancer and chronic inflammation associated with aging.
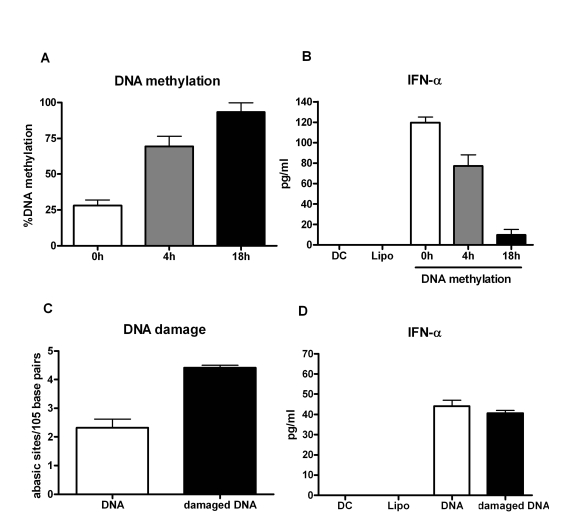
Figure 3. Immunogenicity of mammalian DNA correlates inversely with DNA methylation.
(A) DNA
was methylated using a methyl transferase enzyme and percent global
methylation was measured by ELISA. Bar diagram shows the percent of global
methylation at 0h, 4h and 16h after the reaction. Figure is mean +
S.E. of five separate DNA tested. (B) The immunogenicity of the
methylated DNA was determined by measuring the IFN-α secretion by DCs. Bar diagram
shows the level of IFN-α
secreted by DCs in response to the DNA. Figure is mean +
S.E. of
five separate DNA tested. (C) PBMCs were treated with hydrogen
peroxide to induce DNA damage. Damaged DNA was extracted and the extent of
damage was determined by ELISA. Bar diagram shows the level of DNA damage
before and after treatment. Figure is mean +
S.E. of five separate
DNA tested (D) The immunogenicity of the H2O2
damaged DNA was determined by measuring the IFN-α secretion by DCs. Bar diagram
shows the level of IFN-α
secreted by DCs in response to the DNA. Figure is mean +
S.E. of
five separate DNA tested.
Future
studies of pivotal interest would be the identification of the receptor and
signaling pathways involved in the recognition of this hypomethylated DNA. Earlier studies with intracellular DNA delivered via
transfection reagents have shown that DNA signals through non-TLR receptors [21-23]. Our own study [20] also found that the DNA was localized in the
cytosol and was not accessible to intracellular TLRs in the endosomes. The
nucleic acid sensing TLR3 and TLR8 are found in the endosomes [45]. The two other known nucleic acid sensing TLRs, TLRs 7 and 9 are
not expressed in human monocyte derived DCs and are also present in the
endosomes [46]. Identification of the receptor would also provide novel target
for therapy of autoimmune diseases.
Materials and method
Blood
donors.
Blood was collected from
healthy elderly (age 65-90 years) and young volunteer (age 20-35 years) donors.
Elderly subjects belong to middle-class socio-economic status and are living
independently. A week prior to the study, they were asked to discontinue any
vitamins, minerals and antioxidants that they may have been taking. The
Institutional Review Board of the University of California, Irvine, approved
this study.
Preparation
of human monocyte derived dendritic cells.
DCs were prepared essentially as described [43]. Briefly, peripheral
blood mononuclear cells (PBMCs) were separated by Ficoll-Hypaque density
gradient centrifugation. Monocytes were purified from the PBMCs by positiveselection with anti-CD14 microbeads (Stemcell Sep, Vancouver, BC). The purity
of the isolatedCD14+ monocytes was >90% as determined
by flow cytometry.For the induction of DC differentiation, purified
CD14+ monocytes were cultured in a humidified atmosphere of 5%CO2
at 37°C in RPMI 1640 supplemented with10%FBS, 1 mMglutamine, 100
U/ml penicillin, 100 μg/ml streptomycin,50 ng/ml human rGM-CSF
(PeproTech, Rocky Hill, NJ), and 10 ng/ml human rIL-4(Peprotech).
Half of the medium was replaced every2 days and DCs (CD14-HLA-DR+CD11c+cells) were collected after 6 days. The purity of the DC was >95% as
determined by the expression of CD14, CD11c and HLA-DR.
Self DNA Preparation.
DNA was
isolated from the blood of aged and young subjects using Qiamp DNA Blood Midi
Kit from Qiagen (Valencia, CA). RNase was added to remove any contaminating
RNA. Purity and yield of DNA was measured by UV spectrophotometer. Preparations
with 260/280 ratio above 1.9 were used in all experiments. DNA obtained was
free of endotoxin contamination as determined by Limulus amoebocytelysate
(LAL) assay (Lonza Inc, Allendale, NJ).
DC
activation.
Transfection reagent Lipofectamine 2000 (Invitrogen,
Carlsbad, CA) was used to deliver self DNA to DCs. DNA was mixed with
lipofectamine (lipo) in 100 μl of Opti-MEM for 20 minutes at
room temperature, according to the protocol recommended by the manufacturer and
added to 4×105 DCs in 300 μl of complete
medium. Final concentration of the DNA was 1μg/ml. Cell viability
was unaffected by this treatment. Unstimulated and Lipofectamine-stimulated DCs
were used as controls. After 24 hours, cells were harvested and stained for
surface markers CD80, CD86 and CD83, using directly conjugated antibodies and
isotype controls (BD Pharmingen, San Jose, CA). 10,000 CD11c+HLA-DR+
cells per condition were acquired using a FACSCalibur (BDPharmingen,
San Jose, California). Analysis was performed using the Flow jo software
(Treestar Inc, Ashland, OR).
Cytokine,
IFN-α in the supernatants was measured by verikine IFN-α measuring kit (PBL Biomedicals, Piscatway, NJ) as per the
manufacturer's protocol.
Quantification
of DNA methylation.
Global
methylation of DNA from aged and young subjects was determined using the Methylamp™ Global DNA Methylation Quantification Kit
from Epigentek (Brooklyn, NY), as per the manufacturer's protocol. In this kit
the methylated fraction of DNA is recognized by 5-methylcytosine
antibody and quantified through an ELISA-like reaction.
Methylation
of DNA.
1μg of DNA was methylated in GC Reaction Buffer containing 60 units of
GpC Methyltransferase (M.CviPI) and 160 μM
S-adenosylmethionine from New England Biolabs (Ipswich, MA) at 37oC
for either 4 or 18 hours. The GC Methyltransferase methylates all cytosine
residues within the double stranded recognition sequence of 5'..GC..3'. Percent
methylation was determined using the global DNA methylation kit.
Induction
of DNA damage.
PBMCs from young
donors were treated with 10uM hydrogen peroxide for 1 h at 37oC. DNA
was extracted as described from both treated and untreated PBMCs. Damage was assessed
using the DNA damage quantification kit.
Quantification
of DNA damage.
DNA damage from aged
and young subjects was quantified using DNA damage Quantification kit from
BioVision (Mountain View, CA) following the manufacturer's protocol. This kit
determines the number of abasic sites in purified DNA samples utilizes the ARP
(Aldehyde Reactive Probe) reagent that reacts specifically with an aldehyde
group, which is the open ring form of the Apurinic/apyrimidinic
(
AP) sites.
Statistics.
Statistical analysis was performed using GraphPad
Prism™ 4.00 software (GraphPad Software, San Diego, USA). Unpaired data were
analyzed with the Mann-Whitney test. Wilcoxon signed ranked test was used for
paired analyses. Statistical significance was acknowledged when the P-value
was <0.05.
Acknowledgments
This
study is supported in part by grant AG027512 from NIH and by New Scholar grant
from the Ellison Medical Foundation.
Conflicts of Interest
The authors of this manuscript have no conflict of
interest to declare.
References
-
1.
McGeer
PL
and McGeer
EG.
Inflammation and the degenerative diseases of aging.
Ann N Y Acad Sci.
2004;
1035:
104
-116.
[PubMed]
.
-
2.
Weyand
CM
, Fulbright
JW
and Goronzy
JJ.
Immunosenescence, autoimmunity, and rheumatoid arthritis.
Exp Gerontol.
2003;
38:
833
-841.
[PubMed]
.
-
3.
Boren
E
and Gershwin
ME.
Inflamm-aging: autoimmunity, and the immune-risk phenotype.
Autoimmun Rev.
2004;
3:
401
-406.
[PubMed]
.
-
4.
Ramos-Casals
M
, Garcia-Carrasco
M
, Brito
MP
, Lopez-Soto
A
and Font
J.
Autoimmunity and geriatrics: clinical significance of autoimmune manifestations in the elderly.
Lupus.
2003;
12:
341
-355.
[PubMed]
.
-
5.
Bruunsgaad
H
and Pedersen
BK.
Age-related inflammatory cytokines and disease.
Immunol Allergy Clin North Am.
2003;
23:
15
-39.
[PubMed]
.
-
6.
Ginaldi
L
, De
Martinis M
, Monti
D
and Franceschi
C.
The immune system in the elderly: activation-induced and damage-induced apoptosis.
Immunol Res.
2004;
30:
81
-94.
[PubMed]
.
-
7.
Richardson
BC
Role of DNA methylation in the regulation of cell function: Autoimmunity, aging and cancer.
J Nutr.
2002;
132:
2401S
-2405S.
[PubMed]
.
-
8.
Chen
JH
, Hales
CN
and Ozanne
SE.
DNA damage, cellular senescence and organismal ageing: causal or correlative.
Nucleic Acids Res.
2007;
35:
7417
-7428.
[PubMed]
.
-
9.
Yung
RL
and Julius
A.
Epigenetics, aging, and autoimmunity.
Autoimmunity.
2008;
41:
329
-335.
[PubMed]
.
-
10.
Schumacher
B
, Hoeijmakers
JH
and Garinis
GA.
Sealing the gap between nuclear DNA damage and longevity.
Mol Cell Endocrinol.
2009;
299:
112
-117.
[PubMed]
.
-
11.
Campisi
J
and Vijg
J.
Does damage to DNA and other macromolecules play a role in aging? If so, how.
J Gerontol A Biol Sci Med Sci.
2009;
64:
175
-178.
[PubMed]
.
-
12.
Golbus
J
, Palella
TD
and Richardson
BC.
Quantitative changes in T cell DNA methylation occur during differentiation and ageing.
Eur J Immunol.
1990;
20:
1869
-1872.
[PubMed]
.
-
13.
Issa
JP
Age-related epigenetic changes and the immune system.
Clin Immunol.
2003;
1091:
103
-108.
[PubMed]
.
-
14.
Gaudet
F
, Hodgson
JG
, Eden
A
, Jackson-Grusby
L
, Dausman
J
, Gray
JW
, Leonhardt
H
and Jaenisch
R.
Induction of tumors in mice by genomic hypomethylation.
Science.
2003;
300:
489
-492.
[PubMed]
.
-
15.
Sedelnikova
OA
, Horikawa
I
, Zimonjic
DB
, Popescu
NC
, Bonner
WM
and Barrett
JC.
Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks.
Nat Cell Biol.
2004;
6:
168
-170.
[PubMed]
.
-
16.
Lombard
DB
, Chua
KF
, Mostoslavsky
R
, Franco
S
, Gostissa
M
and Alt
FW.
DNA repair, genome stability, and aging.
Cell.
2005;
120:
497
-512.
[PubMed]
.
-
17.
Martien
S
and Abbadie
C.
Acquisition of oxidative DNA damage during senescence: the first step toward carcinogenesis.
Ann N Y Acad Sci.
2007;
1119:
51
-63.
[PubMed]
.
-
18.
Maynard
S
, Schurman
SH
, Harboe
C
, de Souza-Pinto
NC
and Bohr
VA.
Base excision repair of oxidative DNA damage and association with cancer and aging.
Carcinogenesis.
2009;
30:
2
-10.
[PubMed]
.
-
19.
Rodier
F
, Coppé
JP
, Patil
CK
, Hoeijmakers
WA
, Muñoz
DP
, Raza
SR
, Freund
A
, Campeau
E
, Davalos
AR
and Campisi
J.
Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion.
Nat Cell Biol.
2009;
11:
973
-979.
[PubMed]
.
-
20.
Agrawal
AJ
Tay S, Ton S, Agrawal S, and Gupta S. Increased Reactivity of Dendritic Cells from Aged Subjects to Self Antigen, the Human DNA.
J Immunol.
2009;
182:
1138
-1145.
[PubMed]
.
-
21.
Okabe
Y
, Kawane
K
, Akira
S
, Taniguchi
T
and Nagata
S.
Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation.
J Exp Med.
2005;
202:
1333
-1339.
[PubMed]
.
-
22.
Martin
DA
and Elkon
KB.
Intracellular mammalian DNA stimulates myeloid dendritic cells to produce type I interferons predominantly through a toll-like receptor 9-independent pathway.
Arthritis Rheum.
2006;
54:
951
-962.
[PubMed]
.
-
23.
Ishii
KJ
, Coban
C
, Kato
H
, Takahashi
K
, Torii
Y
, Takeshita
F
, Ludwig
H
, Sutter
G
, Suzuki
K
, Hemmi
H
, Sato
S
and Yamamoto
M.
A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA.
Nat Immunol.
2006;
7:
40
-48.
[PubMed]
.
-
24.
Agrarwal
S
, Gollapudi
S
and Gupta
S.
Increased TNF-alpha-induced apoptosis in lymphocytes from aged humans: changes in the expression of TNF-receptors and activation of caspases.
J Immunol.
1999;
162:
2154
-2161.
[PubMed]
.
-
25.
Garinis
GA
, van der Horst
GTJ
, Vijg
J
and Hoeijmakers
JHJ.
DNA damage and ageing: new-age ideas for an age-old problem.
Nature Cell Biology.
2008;
10:
1241
-1247.
.
-
26.
Romanov
GA
and Vanyushin
BF.
Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction.
Biochim Biophys Acta.
1981;
653:
204
-218.
[PubMed]
.
-
27.
Singhal
RP
, Mays-Hoopes
LL
and Eichhorn
GL.
DNA methylation in aging of mice.
Mech Ageing Dev.
1987;
41:
199
-210.
[PubMed]
.
-
28.
Vanyushin
BF
, Mazin
AL
, Vasilyev
VK
and Belozersky
AN.
The content of 5-methylcytosine in animal DNA: The species and tissue specificity.
Biochim Biophys Acta.
1973;
299:
397
-403.
[PubMed]
.
-
29.
Yano
S
, Ghosh
P
, Kusaba
H
, Buchholz
M
and Longo
DL.
Effect of promoter methylation on the regulation of IFN-gamma gene during in vitro differentiation of human peripheral blood T cells into a Th2 population.
J Immunol.
2003;
171:
2510
-2516.
[PubMed]
.
-
30.
Ballestar
E
, Esteller
M
and Richardson
BC.
The epigenetic face of systemic lupus erythematosus.
J Immunol.
2006;
176:
7143
-147.
[PubMed]
.
-
31.
Sekigawa
I
, Kawasaki
M
, Ogasawara
H
, Kaneda
K
, Kaneko
H
, Takasaki
Y
and Ogawa
H.
DNA methylation: its contribution to systemic lupus erythematosus.
Clin Exp Med.
2006;
6:
99
-106.
[PubMed]
.
-
32.
Ogasawara
H
, Okada
M
, Kaneko
H
, Hishikawa
T
, Sekigawa
I
and Hashimoto
H.
Possible role of DNA hypomethylation in the induction of SLE: relationship to the transcription of human endogenous retroviruses.
Clin Exp Rheumatol.
2003;
21:
733
-738.
[PubMed]
.
-
33.
Wilson
AS
, Power
BE
and Molloy
PL.
DNA hypomethylation and human diseases.
Biochim Biophys Acta.
2007;
1775:
138
-162.
[PubMed]
.
-
34.
Quddus
J
, Johnson
KJ
, Gavalchin
J
, Amento
EP
, Chrisp
CE
, Yung
RL
and Richardson
BC.
Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice.
J Clin Invest.
1993;
92:
38
-53.
[PubMed]
.
-
35.
Wen
ZK
, Xu
W
, Xu
L
, Cao
QH
, Wang
Y
, Chu
YW
and Xiong
SD.
DNA hypomethylation is crucial for apoptotic DNA to induce systemic lupus erythematosus-like autoimmune disease in SLE-non-susceptible mice.
Rheumatology.
2007;
46:
1796
-1803.
[PubMed]
.
-
36.
Issa
JP
, Ottaviano
YL
, Celano
P
, Hamilton
SR
, Davidson
NE
and Baylin
SB.
Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon.
Nat Genet.
1994;
7:
536
-540.
[PubMed]
.
-
37.
Kanai
Y
and Hirohashi
S.
Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state.
Carcinogenesis.
2007;
28:
2434
-2442.
[PubMed]
.
-
38.
Piyathilake
CJ
, Henao
O
, Frost
AR
, Macaluso
M
, Bell
WC
, Johanning
GL
, Heimburger
DC
, Niveleau
A
and Grizzle
WE.
Race- and age-dependent alterations in global methylation of DNA in squamous cell carcinoma of the lung (United States).
Cancer Causes Control.
2003;
14:
37
-42.
[PubMed]
.
-
39.
Post
WS
, Goldschmidt-Clermont
PJ
, Wilhide
CC
, Heldman
AW
, Sussman
MS
, Ouyang
P
, Milliken
EE
and Issa
JP.
Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system.
Cardiovasc Res.
1999;
43:
985
-991.
[PubMed]
.
-
40.
Liu
L
, Wylie
RC
, Andrews
LG
and Tollefsbol
TO.
Aging, cancer and nutrition: The DNA methylation connection.
Mech Ageing Dev.
2003;
124:
989
-998.
[PubMed]
.
-
41.
Stacey
KJ
, Young
GR
, Clark
F
, Sester
DP
, Roberts
TL
, Naik
S
, Sweet
MJ
and Hume
DA.
The molecular basis for the lack of immunostimulatory activity of vertebrate DNA.
J Immunol.
2003;
170:
3614
-3620.
[PubMed]
.
-
42.
Okabe
Y
, Kawane
K
, Akira
S
, Taniguchi
T
and Nagata
S.
Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation.
J Exp Med.
2005;
202:
1333
-1339.
[PubMed]
.
-
43.
Agrawal
A
, Agrawal
S
, Cao
JN
, Su
H
, Osann
K
and Gupta
S.
Altered innate immune functioning of dendritic cells in elderly humans: a role of phosphoinositide 3-kinase-signaling pathway.
J Immunol.
2007;
178:
6912
-6922.
[PubMed]
.
-
44.
Aprahamian
T
, Takemura
Y
, Goukassian
D
and Walsh
K.
Ageing is associated with diminished apoptotic cell clearance in vivo.
Clin Exp Immunol.
2008;
152:
448
-455.
[PubMed]
.
-
45.
Latz
E
, Schoenemeyer
A
, Visintin
A
, Fitzgerald
KA
, Monks
BG
, Knetter
CF
, Lien
E
, Nilsen
NJ
, Espevik
T
and Golenbock
DT.
TLR9 signals after translocating from the ER to CpG DNA in the lysosome.
Nat Immunol.
2004;
5:
190
-198.
[PubMed]
.
-
46.
Ito
T
, Wang
YH
and Liu
YJ.
Plasmacytoid dendritic cell precursors/type I interferon-producing cells sense viral infection by Toll-like receptor (TLR) 7 and TLR9.
Springer Semin Immunopathol.
2005;
26:
221
-229.
[PubMed]
.