Circadian clocks are operative in
virtually all light-sensitive organisms, including cyanobacteria, fungi,
plants, protozoans and metazoans. These timing devices allow their possessors
to adapt their physiological needs to the time of day in an anticipatory way.
In mammals, circadian pacemakers regulate many systemic processes, such as
sleep-wake cycles, body temperature, heartbeat, and many physiological outputs
conducted by peripheral organs, such as liver, kidney and the digestive tract
[1]. On the basis of surgical ablation and transplantation experiments, it was
established that the suprachiasmatic nucleus (SCN) in the hypothalamus
coordinates most of these daily rhythms [2], probably through both synaptic
connections and humoral signals [3]. Interestingly, self-sustained
and cell-autonomous molecular oscillators do not only exist in pacemaker cells
such as SCN neurons, but are also operative in most peripheral, non-neuronal
cell types [4]. These peripheral oscillators participate in the circadian
control
Research Perspective
of
animal physiology. During the past few years, analysis of animal transcriptomes
with the DNA microarray technology showed that many aspects of physiology are
directly controlled by the circadian clock through control of the expression of
enzymes and regulators involved in these physiological processes [5,6].
Although the mechanisms involved in these regulations are not yet understood in
detail, it is likely that transcription factors whose expression is controlled
by the circadian clock are involved [7]. Based on these circadian transcriptome
profiling studies it is commonly thought that circadian metabolism is mainly
the consequence of circadian transcription and possible effects of circadian
clock-controlled post-transcriptional regulatory mechanisms have been largely
neglected.
Interestingly, most of the enzymes involved in liver
metabolism are localized in the membrane of the endoplasmic reticulum (ER) of
hepatocytes. The ER is a complex luminal network in which protein synthesis,
maturation, folding, and transport take place. It has been previously shown
that the ER of hepatocytes exhibits a circadian dilatation which is a sign of
ER stress [8]. This ER stress triggers the unfolded protein response (UPR)
which is a conserved adaptative response to cope with the accumulation of
unfolded proteins in this organelle. When unfolded proteins accumulate in ER,
three pathways are activated, IRE1α,
PERK and ATF6, which lead to the nuclear translocation of the transcription
factors XBP1, ATF4 and ATF6, respectively. These transcription factors activate
in turn the expression of genes coding for proteins involved in peptide folding
and degradation to limit the accumulation of unfolded proteins [9]. In this context, we have recently described the
posttranslational regulation of liver enzymes through a circadian
clock-coordinated 12-hours period rhythmic activation of the IRE1α pathway [10]. The observed rhythmic activation of the
IRE1α pathway leads to the expression with a 12-hours period
of the XBP1-regulated genes that are included in the 12-hours period genes
described recently in mouse liver [11]. Persistent activation of the IRE1α pathway in circadian clock deficient Cry1/Cry2
ko mice induced the downregulation of ER membrane localized enzymes, including
HMGCR and SCD1, leading to a perturbed lipid metabolism in the liver of this
mice. The decreased expression of these enzymes could be caused by activation
of the ER Associated Degradation (ERAD), a process involved in the elimination of unfolded
proteins inside the ER[12]
regulated by the IRE1α-XBP1 pathway [13],
which has been shown to induce the degradation of HMGCR and SCD1. In addition, IRE1α is a ribonuclease that can also induce
endonucleolytic decay of many ER-localized mRNA including Hmgcr mRNA
[14,15]. These two functions could contribute in parallel to the regulation of
lipid metabolism by ER stress. Elsewhere, the IRE1α-XBP1 pathway controls also lipid metabo-lism through
direct transcriptional regulation of the genes Scd1, Dgat2 and Acc2
involved in lipogenesis. As a consequence, liver-specific deletion of the Xpb1
gene resulted in a dramatic reduction of plasma lipids [16]. Finally, it has
been shown that ER stress induces the degradation of the apolipoprotein ApoB100
[17,18] and then blocks VLDL secretion [19], which might be responsible for the
fat accumulation in the liver in tunicamycin-injected mice [20].
Interestingly, IRE1α activation has been recently linked to induction of
autophagy through activation of the Jun-Kinase pathway [21]. In addition, a
genomic screen in fly cells demonstrated that knocking down genes involved in
protein folding inside the ER or in the UPR, including Xbp1, increases
basal autophagy levels [22]. Autophagy is a survival pathway classically
associated with adaptation to nutrient starvation [23] and, as UPR, autophagy
presented a diurnal rhythm of activation in rodent liver [24,25]. This is of
particular interest if we consider the fact that autophagy is linked to lipid
metabolism through regulation of intracellular lipid stores [26]. As a
consequence, mice with an adipose tissue-specific deletion of the Atg7 gene,
an important regulator of autophagy, present an important defect in lipid
storage [27,28]. IRE1α-dependent rhythmic
regulation of autophagy could then participates to the circadian
clock-coordinated lipid metabolism in mammals.
The disturbed metabolism observed in Cry1/Cry2
ko mice is probably responsible of the aberrant activation of the Sterol Responsive Element Binding Protein (SREBP)
transcription factor, an ER membrane bond protein that, in low sterol
conditions, translocates to the Golgi to be cleaved and released in order to
migrate in the nucleus where it activates genes coding for enzymes involved in
cholesterol and fatty acid metabolism [29]. It has been shown that the ER
stress induced activation of SREBP1 and SREBP2 [30,31] correlates with the
depletion of INSIG regulatory proteins
probably through a decreased synthesis of the protein [32,33]. Interestingly, the circadian clock influences
also the activation of the SREBP pathway through the control of Insig2
mRNA expression [34]. Both transcriptional and post-transcriptional circadian
clock-coordinated events seem to be involved in the rhythmic activation of the
SREBP pathway.
As summarised in Figure 1, in addition to their rhythmic activation, all these pathways have in common the fact that they are
regulated by feeding-fasting events. However, this feeding rhythm, like most
behaviour, is also controlled by the circadian clock. To discriminate the genes
dependant or not on a functional local circadian oscillator, this local clock
has been inactivated in mouse liver. This strategy reveals that the expression
of approximately 90 % of the rhythmic genes is dependent on a functional
circadian clock and only 10 % is dependent on systemic cues [35]. However, the
influence of feeding on rhythmic gene expression has been evaluated by a recent
study which discriminates between gene induced by feeding and fasting. As
expected, food-induced and food-repressed genes present a rhythmic expression
which is shifted in response to a change in the feeding schedule [36]. More
interestingly, this shift in the feeding schedule is able to induce rhythmic
expression of food-regulated genes in the liver of Cry1/Cry2
ko mice. These two studies raise the
question of the differential influence of the molecular circadian oscillator
and systemic cues on rhythmic gene expression: if these two signals can
independently drive rhythmic gene expression, the circadian clock is able to
fine-tune and modify feeding cues [34,36], whereas feeding cues can synchronize
the molecular oscillator in peripheral organs [37].
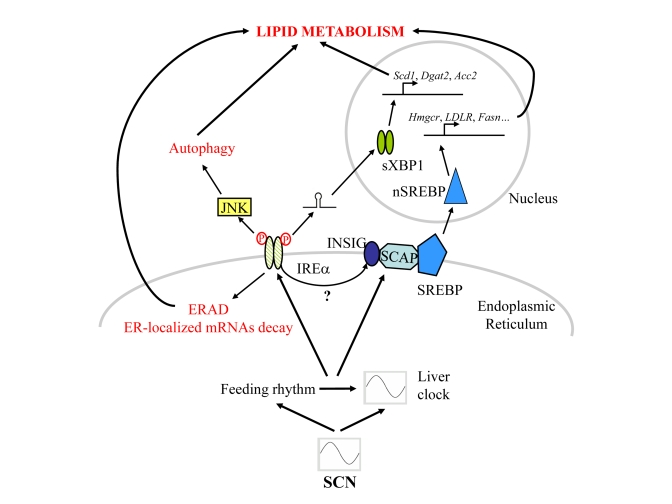
Figure 1. Schematic representation of the signalling pathways post-transcriptionally regulated by the circadian clock and/or rhythmic feeding cues in mouse liver.
However, feeding and food-regulated signals, as for
example food regulated hormones like insulin, glucagon or leptin, did not
represent the only circadian clock-regulated cues that can influence lipid
metabolism. For example, the pituitary-secreted growth hormone (GH) has been
shown to influence lipid metabolism in mouse liver. Long term excess GH
secretion produces high serum triglyceride levels through stimulated lipolysis
[38], whereas inhibition of GH signaling induced perturbed lipid metabolism
resulting in liver steatosis [39], probably caused by reduced activation of
HNF3β [40].
Moreover, the ultradian secretion patterns of GH are directly responsible for
the sexually dimorphic expres-sion of several hepatic enzymes involved in
steroids and fatty acids metabolism [41]. Interestingly, this dimorphism is
impaired in Cry1/Cry2 ko mice, with males exhibiting a feminized
liver likely because of altered ultradian GH secretion in absence of a functional
circadian clock [42].
During
aging, the circadian system becomes much less responsive to entrainment by
light [43,44], and displays loss of temporal precision and robustness [45-47]. Such
alterations of the circadian clock likely drive attenuation of the diurnal
rhythm in circulating leptin [48]. Pulsatile GH secretion is also dramatically
impaired in elderly subjects [49-51], leading to modifications in GH-dependent
liver metabolism that resemble those observed in clock-deficient animals
[42,52]. Interesting-ly, the
various UPR pathways also decline in the liver during aging [53], as well as
autophagy [54]. In summary, many aspects of lipid metabolism that are regulated
by the circadian clock exhibit profound changes when age increases, although
the liver circadian oscillator appears preserved in aged rats [55]. These
changes could thus at least partly originate from alterations of the network
constituted of the central clock and other peripheral oscillators. In this
respect, it is worth noting that mice bearing mutated alleles of the circadian
genes Clock and Bmal1 display signs of premature aging [56,57]. The complexity of systemic cues influencing rhythmic
gene expression has thus been rising during the last decade and defining the
influence of these different signals on rhythmic gene expression will be thus
an exciting challenge for the following years.
The authors of this manuscript have no conflict of interest
to declare.