The sleep-feeding conflict: Understanding behavioral integration through genetic analysis in Drosophila
Abstract
One of the brain's most important functions is the control of homeostatically regulated behaviors. Dysregulation of the neural systems controlling sleep and feeding underlies many chronic illnesses. In a recent study published inCurrent Biology we showed that flies, like mammals, suppress sleep when starved and identified the genes Clock and cycle as regulators of sleep during starvation. Here we show that starvation specifically disrupts sleep initiation without affecting sleep consolidation. The identification of genes regulating sleep-feeding interactions will provide insight into how the brain integrates and controls the expression of complex behaviors.
Sleep
and feeding are mutually exclusive behaviors. Consequently, an animal must
decide which behavior to express based on internal drives and environmental
cues. These behaviors are also functionally interconnected: food-deprivation
suppresses sleep, while sleep loss induces hunger [1,2]. Extreme dysregulation of either
behavior on its own is deleterious. Longitudinal studies in humans have
revealed increased Body Mass Index in short sleeping individuals [3]. The
neuropeptides Orexin and neuropeptide Y (NPY) both suppress sleep and promote
feeding [4,5], while
mice mutant for the leptin receptor have disrupted sleep patterns [6].
Sleep
loss potently affects insulin function and has been clinically linked to Diabetes
mellitus,
metabolic syndromes,
like Diabetes
mellitus
and
obesity. It is possible that the
interplay between sleep and metabolic syndromes occurs
through the direct effect of sleep on metabolism or
indirectly through the dysregulation of appetite [7].
Understanding the molecular and neural link between sleep and feeding will aid in
our
understanding of obesity and sleep-linked disorders.
Much of the genetic architecture
controlling sleep, feeding and metabolism is conserved across phyla. A
powerful
genetic toolkit has been developed in the fruit fly, Drosophila
melanogaster, that allows for the manipulation of genes and neural circuits
with regional and temporal specificity [8]. Genetic
screens in Drosophila have led to the identification of many genes
affecting sleep, feeding and metabolism with conserved function in mammals.
For example the Dopamine transporter promotes sleep [9,10] and a
genome-wide obesity screen identified the hedgehog pathway as a conserved
determinant of fat generation [11].
To
gain insight into the genetic and neural basis of sleep-feeding interactions we
investigated the effects of food-deprivation on Drosophila sleep. Energy
stores and sleep needs are linked suggesting a link between metabolism and
sleep [12]. In
addition, because starved flies only survive 1-2 days, we reasoned they might
be particularly sensitive to the sleep-suppressing effects of food-deprivation.
We
therefore monitored flies' activity over a 24-hour period in small tubes with
either standard fly food or agar as a feeding substrate (Figure 1A, B). We found that wild-type flies robustly suppress
sleep following 12-hours of starvation on agar (Figure 1C, D), suggesting that
the effect of food-deprivation on sleep that was previously documented in
mammals is conserved in Drosophila.
Mammalian sleep is composed of distinct stages that can
be characterized by unique electrophysiological properties. Sleep in flies is a
also accompanied by alterations in neural activity [13], yet the relevance of these
changes to mammalian sleep states remains unclear. Consolidation of sleep can
be measured behaviorally in flies by determining the average length and total
number of individual sleep bouts. Disruption in bout number suggests difficulty
in initiating sleep while shortened bout length indicates a failure to maintain
sleep. We found that 24 hours of starvation decreases bout number without affecting bout length (Figure 1E, F). Therefore,
food-deprivation specifically affects the onset of sleep without affecting
sleep maintenance.
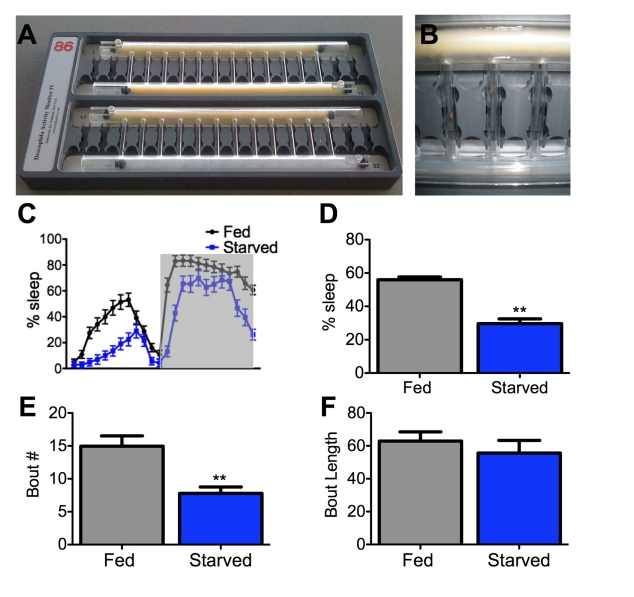
Figure 1. Starvation impairs sleep initiation but not maintenance. (A,B)
A Drosophila activity monitor typically used for sleep studies
can record up to 32 flies simultaneously. An individual fly is housed in
each vertical tube and an infrared beam detects activity. The large
horizontal tubes contain either food (yellow) or agar (translucent).
Sliding barriers control access to each substrate [32]. Both tubes
contain food for fed controls (A, top), while agar is provided to
the starved experimental group (A, bottom; and B) on day 2 of testing the
experiment (starved, experimental). (C,D) Female flies
starved for 24 hours sleep less than fed counterparts. Shaded area (C)
represents lights-off. (E, F) The total number of sleep
bouts (Bout #) is decreased in starved flies while average bout length does
not differ from fed counterparts. Asterisk denotes significant difference
(P<0.01, ANOVA) from control groups. Data are mean ± SEM.
We
screened for mutants with aberrant sleep during starvation in order to identify
genes linking sleep and feeding. We found that mutants for the genes Clock
and cycle are hypersensitive to the wake-promoting effects of
food-deprivation. Clock and cycle are transcriptional activators
that are expressed in ~150 central brain neurons, multiple populations of
sensory neurons and peripheral cells. Clock and cycle function as
binding partners and are required for 24-hour transcriptional cycling of the
core-circadian clock [14].
In
addition to regulating circadian rhythms, Clock and cycle have
been implicated in the regulation of sleep, feeding, olfaction, and starvation
resistance [15-18]. Clock-dependent
modulation of each behavior appears to be conferred through distinct neuronal
populations. For example, Clock-regulated control of circadian behavior
localizes to eight neurons termed the small ventrolateral neurons [15] while
regulation of feeding and starvation resistance localize to the gustatory
neurons and fat body
bodies
[16,18].
Through tissue-specific disruption of Clock function
we probed populations of cells for
their role in starvation-induced sleep suppression. Selectively disrupting Clock
function in a population of dorsally located neurons in the central brain
phenocopied the genetic mutant. However, eliminating Clock function in
cells previously implicated in circadian locomotor behavior, feeding,
olfaction, vision, or starvation-resistance did not affect sleep-suppression
during starvation. Therefore, cellular control of sleep-feeding interactions
appears to be distinct from those controlling other Clock-dependent
behaviors.
The
pleiotropic nature of behavior suggests many additional genes function in
concert with Clock and cycle to modulate sleep-feeding
interactions. Neuropeptide F, the Drosophila ortholog of Neuropeptide
Y, has been implicated in control of feeding [19] and
motivational behavior [20] and is an
excellent candidate for modulating sleep-feeding interactions. The mammalian
gastrointestinal satiety-inducing peptide cholecys
tokinin (CCK) has been reported to induce sleep and a CCK-A
receptor antagonist blocks
this effect [21]. The function
of drosulfakinin, the fly ortholog of CCK, is unknown. It is expressed
in the brain [22] and
represents a candidate for signaling nutrients
cues to Clock-expressing neurons.
In
mammals, hypothalamic Orexin regulates both sleep and feeding and Orexin
signaling has been proposed as an attractive drug target for dysfunction of
both sleep and feeding systems [23,24]. In
addition to Orexin, T-type Ca2+ channels have been linked to
regulation of sleep-feeding interactions. Administration of a selective T-Type
Ca2+ channel antagonist increases sleep and reduces body fat in mice
fed a high-fat diet [25]. In flies,
Ca2+ homeostasis has been linked to sleep-wake regulation [26] and future
investigation of the role of specific Ca2+ channels in the
regulation of sleep and feeding may be informative.
Our study focused on the effect of
food-deprivation on sleep. The consequences of sleep-deprivation on metabolism
were not addressed. Loss of sleep has detrimental
effects on metabolism and has been linked to conditions such as obesity and
diabetes [27],
and the Drosophila insulin-producing cells have
been shown to regulate sleep. Hyperexcitation of insulin
insulin-
producing cells inhibits sleep [28] while activation of
the
Epidermal Growth Factor Receptor activation
in
these cells induces sleep [29]. These findings suggest a functional link between
the systems controlling insulin and sleep. Furthermore, alterations in
mice
mutant for
Clock and BMAL1, the mammalian orthologs
of Clock and cycle, have significant metabolic defects that
include decreased insulin release and a
diminished
ability to maintain normal blood glucose levels [30,31]. Future work examining the
metabolism of short-sleeping Drosophila
mutants may aid our understanding
of the link between sleep loss and metabolic dysfunction.
Identifying the molecular basis of behavioral integration will pave the
way for the development of drugs that act in a context-dependent fashion. Our
findings
that Clock and cycle regulate sleep during food-deprivation is a starting point for
understanding the complex interactions regulating sleep and feeding. Utilizing
currently available fly mutants to verify candidate genes identified in
large-scale fly and mammalian analyses should significantly improve our
understanding of sleep-feeding interactions and resulting pathologies.
References
-
1.
MacFadyen
UM
, Oswald
I
and Lewis
SA.
Starvation and human slow-wave sleep.
J Appl Physiol.
1973;
35:
391
-394.
[PubMed]
.
-
2.
Spiegel
K
, Tasali
E
, Penev
P
and Van
Cauter E.
Brief communication: Sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite.
Ann Intern Med.
2004;
141:
846
-850.
[PubMed]
.
-
3.
Taheri
S
, Lin
L
, Austin
D
, Young
T
and Mignot
E.
Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index.
PLoS Med.
2004;
1:
e62
[PubMed]
.
-
4.
Szentirmai
E
and Krueger
JM.
Central administration of neuropeptide Y induces wakefulness in rats.
Am J Physiol Regul Integr Comp Physiol.
2006;
291:
R473
-480.
[PubMed]
.
-
5.
Chemelli
RM
, Willie
JT
, Sinton
CM
, Elmquist
JK
, Scammell
T
, Lee
C
, Richardson
JA
, Williams
SC
, Xiong
Y
and Kisanuki
Y.
Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation.
Cell.
1999;
98:
437
-451.
[PubMed]
.
-
6.
Laposky
AD
, Bradley
MA
, Williams
DL
, Bass
J
and Turek
FW.
Sleep-wake regulation is altered in leptin-resistant (db/db) genetically obese and diabetic mice.
Am J Physiol Regul Integr Comp Physiol.
2008;
295:
R2059
-2066.
[PubMed]
.
-
7.
Spiegel
K
, Knutson
K
, Leproult
R
, Tasali
E
and Van
Cauter E.
Sleep loss: a novel risk factor for insulin resistance and Type 2 diabetes.
J Appl Physiol.
2005;
99:
2008
-2019.
[PubMed]
.
-
8.
Venken
KJ
and Bellen
HJ.
Emerging technologies for gene manipulation in Drosophila melanogaster.
Nat Rev Genet.
2005;
6:
167
-178.
[PubMed]
.
-
9.
Kume
K
, Kume
S
, Park
SK
, Hirsh
J
and Jackson
FR.
Dopamine is a regulator of arousal in the fruit fly.
J Neurosci.
2005;
25:
7377
-7384.
[PubMed]
.
-
10.
Wu
MN
, Koh
K
, Yue
Z
, Joiner
WJ
and Sehgal
A.
A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila.
Sleep.
2008;
31:
465
-472.
[PubMed]
.
-
11.
Pospisilik
JA
, Knauf
C
, Joza
N
, Benit
P
, Orthofer
M
, Cani
PD
, Ebersberger
I
, Nakashima
T
, Sarao
R
and Neely
G.
Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes.
Cell.
2007;
131:
476
-491.
[PubMed]
.
-
12.
Harbison
ST
and Sehgal
A.
Quantitative genetic analysis of sleep in Drosophila melanogaster.
Genetics.
2008;
178:
2341
-2360.
[PubMed]
.
-
13.
Nitz
DA
, van
Swinderen B
, Tononi
G
, Greenspan
RJ: Electrophysiological correlates of rest
and activity
in Drosophila melanogaster.
Curr Biol.
2002;
12:
1934
-1940.
[PubMed]
.
-
14.
Rutila
JE
, Suri
V
, Le
M
, So
WV
, Rosbash
M
and Hall
JC.
CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless.
Cell.
1998;
93:
805
-814.
[PubMed]
.
-
15.
Tanoue
S
, Krishnan
P
, Krishnan
B
, Dryer
SE
and Hardin
PE.
Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila.
Curr Biol.
2004;
14:
638
-649.
[PubMed]
.
-
16.
Chatterjee
A
, Tanoue
S
, Houl
JH
and Hardin
PE.
Regulation of Gustatory Physiology and Appetitive Behavior by the Drosophila Circadian Clock.
Curr Biol.
2010;
20:
300
-309.
[PubMed]
.
-
17.
Hendricks
JC
, Lu
S
, Kume
K
, Yin
JC
, Yang
Z
and Sehgal
A.
Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster.
J Biol Rhythms.
2003;
18:
12
-25.
[PubMed]
.
-
18.
Xu
K
, Zheng
X
and Sehgal
A.
Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila.
Cell Metab.
2008;
8:
289
-300.
[PubMed]
.
-
19.
Wu
Q
, Zhao
Z
and Shen
P.
Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems.
Nat Neurosci.
2005;
8:
1350
-1355.
[PubMed]
.
-
20.
Krashes
MJ
, DasGupta
S
, Vreede
A
, White
B
, Armstrong
JD
and Waddell
S.
A neural circuit mechanism integrating motivational state with memory expression in Drosophila.
Cell.
2009;
139:
416
-427.
[PubMed]
.
-
21.
Shemyakin
A
and Kapas
L.
L-364,718, a cholecystokinin-A receptor antagonist, suppresses feeding-induced sleep in rats.
Am J Physiol Regul Integr Comp Physiol.
2001;
280:
R1420
-1426.
[PubMed]
.
-
22.
Nichols
R
and Lim
IA.
Spatial and temporal immunocytochemical analysis of drosulfakinin (Dsk) gene products in the Drosophila melanogaster central nervous system.
Cell Tissue Res.
1996;
283:
107
-116.
[PubMed]
.
-
23.
Willie
JT
, Chemelli
RM
, Sinton
CM
and Yanagisawa
M.
To eat or to sleep? Orexin in the regulation of feeding and wakefulness.
Annu Rev Neurosci.
2001;
24:
429
-458.
[PubMed]
.
-
24.
Hungs
M
and Mignot
E.
Hypocretin/orexin, sleep and narcolepsy.
Bioessays.
2001;
23:
397
-408.
[PubMed]
.
-
25.
Uebele
VN
, Gotter
AL
, Nuss
CE
, Kraus
RL
, Doran
SM
, Garson
SL
, Reiss
DR
, Li
Y
, Barrow
JC
and Reger
TS.
Antagonism of T-type calcium channels inhibits high-fat diet-induced weight gain in mice.
J Clin Invest.
2009;
119:
1659
-1667.
[PubMed]
.
-
26.
Zimmerman
JE
, Rizzo
W
, Shockley
KR
, Raizen
DM
, Naidoo
N
, Mackiewicz
M
, Churchill
GA
and Pack
AI.
Multiple mechanisms limit the duration of wakefulness in Drosophila brain.
Physiol Genomics.
2006;
27:
337
-350.
[PubMed]
.
-
27.
Knutson
KL
, Spiegel
K
, Penev
P
and Van
Cauter E.
The metabolic consequences of sleep deprivation.
Sleep Med Rev.
2007;
11:
163
-178.
[PubMed]
.
-
28.
Crocker
A
, Shahidullah
M
, Levitan
IB
and Sehgal
A: Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior.
Neuron.
2010;
65:
670
-681.
[PubMed]
.
-
29.
Foltenyi
K
, Greenspan
RJ
and Newport
JW.
Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila.
Nat Neurosci.
2007;
10:
1160
-1167.
[PubMed]
.
-
30.
Marcheva
B
, Ramsey
KM
, Buhr
ED
, Kobayashi
Y
, Su
H
, Ko
CH
, Ivanova
G
, Omura
C
, Mo
S
and Vitaterna
MH.
Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes.
Nature.
2010;
466:
627
-631.
[PubMed]
.
-
31.
Froy
O
and Miskin
R.
Effect of feeding regimens on circadian rhythms: implications for aging and longevity.
Aging.
2010;
2:
7
-27.
[PubMed]
.
-
32.
Agosto
J
, Choi
JC
, Parisky
KM
, Stilwell
G
, Rosbash
M
and Griffith
LC.
Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila.
Nat Neurosci.
2008;
11:
354
-359.
[PubMed]
.