XPG rs17655 G>C polymorphism associated with cancer risk: evidence from 60 studies
Abstract
Xeroderma pigmentosum group G (XPG), a key component in nucleotide excision repair pathway, functions to cut DNA lesions during DNA repair. Genetic variations that alter DNA repair gene expression or function may decrease DNA repair ability and impair genome integrity, thereby predisposing to cancer. The association between XPG rs17655 G>C polymorphism and cancer risk has been investigated extensively, but the results remain contradictory. To get a more accurate conclusion, we performed a comprehensive meta-analysis of 60 case-control studies, involving 27,098 cancer cases and 30,535 healthy controls. Crude odds ratios (ORs) and 95% confidence interval (CIs) were calculated to determine the association of interest. Pooled analysis indicated that the XPG rs17655 G>C polymorphism increased the risk of overall cancer (CC vs. GG: OR=1.10, 95% CI=1.00-1.20; CG vs. GG: OR=1.06, 95% CI=1.02-1.11; CG+CC vs. GG: OR=1.07, 95% CI=1.02-1.12; C vs. G: OR=1.05, 95% CI=1.01-1.09). Stratification analysis by cancer type further showed that this polymorphism was associated with increased risk of gastric cancer and colorectal cancer. This meta-analysis indicated that the XPG gene rs17655 G>C polymorphism was associated with increased overall cancer risk, especially the risk of gastric cancer and colorectal cancer. Further validation experiments are needed to strength our conclusion.
Introduction
Cancer-related deaths continue to rise in both developed and developing countries. In 2012, there were about 14.1 million new cancer cases and 8.2 million cancer-related deaths all over the world. Lung and breast cancer are the most common forms of cancer in human beings. Moreover, the incidences of liver, stomach and colorectal cancer are also very high in men and stomach, while cervix uteri and colorectal cancer prevail in women. Cancer is a complex disease. A variety of cancer risk factors have been recognized, such as smoking, drinking, lack of exercise, poor diet, reproductive changes, and genetic lesions [1]. Inherited genetic causations of cancer risk are mainly unidentified. Thus far, great effects have been made to discover genetic variant alleles implicated in the crucial signaling pathways, which may influence individual cancer predisposition.
Genetic DNAs of living organisms are constantly subjected to various types of damages caused by environmental agents and byproducts (e.g., reactive oxygen species) of cellular metabolic processes. To maintain genome integrity, human beings possess a number of systems for the prevention and restoration of DNA damage. Reduced DNA repair ability is a predisposing factor to cancer [2]. Five common DNA repair pathways have been identified, including nucleotide excision repair (NER), base excision repair, double-strand DNA break repair, mismatch repair, and transcription coupled repair [3,4]. Among these pathways, NER is responsible for removing damaged DNA fragments (e.g., bulky adducts) resulting from radiation or chemical agents [5,6]. In the NER pathway, at least eight vital genes [excision repair cross-complementation group 1 (ERCC1), ERCC2/ Xeroderma pigmentosum group D (XPD), ERCC3/XPB, ERCC4/XPF, ERCC5/XPG, XPA, XPC and XPE/damaged DNA-binding protein 1 (DDB1)] have been well studied, which participate in DNA repair, capable of preserving genetic integrity to prevent cells from malignant transformation [7].
ERCC5/XPG is located on chromosome 13q22-33, consisting of 15 exons and 14 introns . Its protein product is a 1,186 amino acid structure-specific endonuclease, and plays an essential role in the two incision steps of NER [4,8]. XPG is highly polymorphic. Among known single nucleotide polymorphisms (SNPs) in this gene, a nonsynonymous Asp1104His (rs17655, G>C) polymorphism is most frequently studied for its association with cancer risk [2,9–38]. However the results are inconsistent from study to study. Therefore, we performed this meta-analysis with all eligible publications to investigate the association between the XPG gene rs17655 G>C polymorphism and cancer risk.
Results
Study characteristics
As shown in Figure 1, we found 362 potentially relevant studies from PubMed, EMBASE, CNKI, WANFANG, and Vip databases. After reviewing titles and abstracts, we excluded 281 publications not investigating the association between XPG gene rs17655 polymorphism and cancer risk. And then, full texts of remaining articles were evaluated. Two publications [39,40] were removed for containing overlap data. We also excluded 11 publications [41–51] because no sufficient data were reported to calculate ORs and 95% CIs. Furthermore, we eliminated five publications [52–56] presenting survival data only. At last, we excluded five publications [57–61] due to deviation from HWE. In the end, 58 publications with a total of 27,098 cancer cases and 30,535 healthy controls were included in the meta-analysis. It was noteworthy that, 58 publications actually consisted of 60 case-control studies, because 2 of them included two individual studies. The characteristics of these studies were showed in Table 1. Among these publications, five focused on gastric cancer [15,22,31,37,38], 10 on breast cancer [18,29,33,34,59,62–66], four on colorectal cancer [16,20,25,67], four on lymphoma [11,21,68,69], six on bladder cancer [24,70–74], five on lung cancer [17,30,75–77], eight on skin cancer [14,23,26,32,35,78–80], three on HNC [10,81,82], two on endometrial cancer [19,83], laryngeal carcinoma [9,84], and prostate cancer [12,28]. Moreover, there was only one study for each of the following cancers: osteosarcoma [13], hepatocellular carcinoma [36], esophageal carcinoma [85], oral squamous cell carcinoma [86], sarcoma [2], cervical carcinoma [27] and brain cancer [87]. Among these case-control studies, 25 of them had quality scores higher than 9, while 35 had quality scores no more than 9. Finally, this meta-analysis contained 26 hospital-based, 31 population-based, and three mixed control studies.
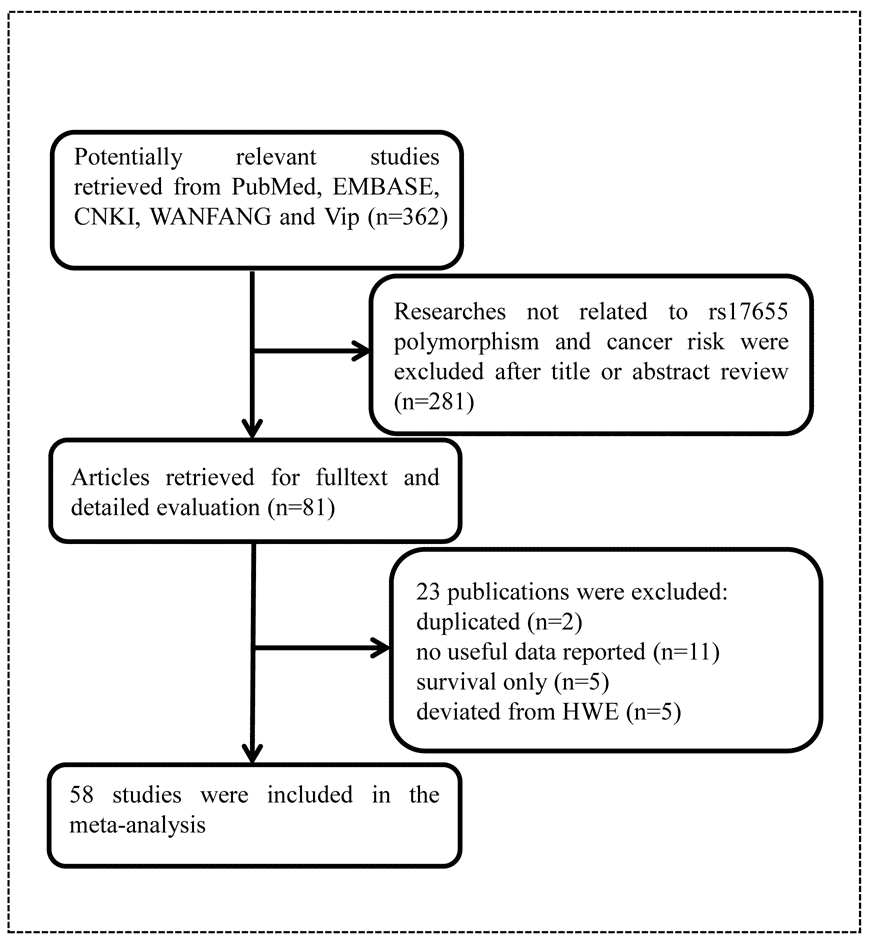
Figure 1. Flowchart of included publications.
Table 1. Characteristics of included studies in the final meta-analysis.
| Name | Year | Cancer type | Region | Ethnicity | Design | Genotype | Case | Control | MAF | HWE | Score |
| | | | | | method | GG | CG | CC | All | GG | CG | CC | All | | | |
| Feng | 2016 | Gastric | China | Asian | HB | PCR-RFLP | 47 | 85 | 45 | 177 | 84 | 107 | 46 | 237 | 0.42 | 0.260 | 6 |
| Ma | 2016 | Breast | China | Asian | HB | PCR-RFLP | 116 | 145 | 59 | 320 | 84 | 107 | 46 | 237 | 0.42 | 0.260 | 7 |
| Du | 2016 | Colorectal | China | Asian | HB | TaqMan | 286 | 459 | 133 | 878 | 355 | 405 | 124 | 884 | 0.37 | 0.623 | 9 |
| Wang | 2015 | Breast | China | Asian | HB | PCR-RFLP | 95 | 6 | 0 | 101 | 100 | 1 | 0 | 101 | 0.00 | 0.960 | 9 |
| Bahceci | 2014 | B-NHL | Turkey | Others | PB | AS-PCR | 59 | 33 | 1 | 93 | 43 | 44 | 9 | 96 | 0.32 | 0.637 | 4 |
| Li | 2014 | Gastric | China | Asian | HB | PCR-RFLP | 99 | 83 | 36 | 218 | 112 | 82 | 24 | 218 | 0.30 | 0.135 | 7 |
| Zhu | 2014 | Bladder | China | Asian | HB | MassARRAY | 62 | 160 | 65 | 287 | 76 | 139 | 67 | 282 | 0.48 | 0.825 | 6 |
| Lu | 2014 | Larynx | China | Asian | HB | MassARRAY | 53 | 69 | 54 | 176 | 78 | 63 | 36 | 177 | 0.38 | 0.001 | 8 |
| Liu | 2014 | Gastric | China | Asian | HB | PCR-RFLP | 99 | 100 | 39 | 238 | 120 | 95 | 23 | 238 | 0.30 | 0.510 | 8 |
| Ruiz-Cosano | 2013 | BCL | Spain | Caucasian | PB | TaqMan | 125 | 71 | 17 | 213 | 119 | 81 | 14 | 214 | 0.25 | 0.965 | 7 |
| Zeng | 2013 | Lung | China | Asian | HB | PCR-RFLP | 15 | 77 | 47 | 139 | 35 | 61 | 37 | 133 | 0.51 | 0.341 | 8 |
| Yuan | 2012 | HNC | China | Asian | PB | TaqMan | 108 | 191 | 95 | 393 | 234 | 433 | 217 | 884 | 0.49 | 0.552 | 12 |
| Biason | 2012 | Osteosarcoma | Italy | Caucasian | HB | PCR-RFLP | 75 | 39 | 16 | 130 | 141 | 94 | 15 | 250 | 0.25 | 0.899 | 8 |
| Gil | 2012 | Colorectal | Poland | Caucasian | HB | PCR-RFLP | 86 | 35 | 11 | 132 | 64 | 31 | 5 | 100 | 0.21 | 0.625 | 6 |
| Berhane | 2012 | Prostate | India | Asian | PB | PCR-RFLP | 58 | 72 | 20 | 150 | 66 | 75 | 9 | 150 | 0.31 | 0.039 | 8 |
| Ma | 2012 | HNC | America | Caucasian | PB | SNPlex | 648 | 359 | 52 | 1059 | 654 | 350 | 62 | 1066 | 0.22 | 0.099 | 10 |
| Rouissi | 2011 | Bladder | Tunisia | African | PB | PCR | 48 | 56 | 21 | 125 | 46 | 61 | 18 | 125 | 0.39 | 0.758 | 6 |
| Ibarrola-Villava | 2011 | Melanoma | Spain | Caucasian | HB | TaqMan | 326 | 222 | 50 | 598 | 215 | 140 | 24 | 379 | 0.25 | 0.85 | 5 |
| Canbay | 2011 | Colorectal | Turkey | Others | PB | PCR-RFLP | 43 | 34 | 2 | 79 | 148 | 83 | 16 | 247 | 0.23 | 0.352 | 10 |
| Goncalves | 2011 | Melanoma | Brazil | Caucasian | HB | PCR-RFLP | 105 | 77 | 10 | 192 | 109 | 74 | 25 | 208 | 0.30 | 0.031 | 9 |
| Doherty | 2011 | Endometrial | America | Others | PB | Unknown | 418 | 254 | 42 | 714 | 408 | 248 | 47 | 703 | 0.24 | 0.268 | 10 |
| Hsu | 2010 | Breast | China | Asian | HB | TaqMan | 76 | 191 | 134 | 401 | 129 | 243 | 159 | 531 | 0.53 | 0.059 | 8 |
| Figl | 2010 | Melanoma | German, Spain | Caucasian | PB | TaqMan | 703 | 409 | 74 | 1186 | 725 | 465 | 84 | 1274 | 0.25 | 0.420 | 8 |
| Canbay | 2010 | Gastric | Turkey | Others | PB | PCR-RFLP | 25 | 12 | 3 | 40 | 148 | 83 | 16 | 247 | 0.23 | 0.352 | 8 |
| Li | 2010 | HCC | China | Asian | HB | TaqMan | 174 | 233 | 93 | 500 | 151 | 265 | 91 | 507 | 0.44 | 0.175 | 11 |
| Narter | 2009 | Bladder | Turkey | Others | PB | PCR-RFLP | 25 | 28 | 3 | 56 | 18 | 19 | 3 | 40 | 0.31 | 0.505 | 5 |
| Abbasi | 2009 | Larynx | Germany | Caucasian | PB | Real-time PCR | 137 | 103 | 8 | 248 | 380 | 230 | 37 | 647 | 0.23 | 0.778 | 11 |
| Hussain | 2009 | Gastric | China | Asian | PB | SNPlex | 38 | 105 | 38 | 181 | 90 | 180 | 90 | 360 | 0.50 | 1.000 | 12 |
| El-Zein | 2009 | HD | America | Caucasian | PB | TaqMan | 104 | 78 | 16 | 198 | 127 | 80 | 12 | 219 | 0.24 | 0.897 | 10 |
| McKean-Cowdin | 2009 | Brain | America | Caucasian | Mixed | TaqMan and MassARRAY | 499 | 348 | 157 | 1004 | 989 | 657 | 311 | 1957 | 0.33 | 0.000 | 13 |
| Pan | 2009 | Esophageal | America | Caucasian | HB | TaqMan | 201 | 131 | 12 | 344 | 287 | 155 | 15 | 457 | 0.20 | 0.281 | 7 |
| Rajaraman | 2008 | Breast | America | Others | PB | TaqMan | 482 | 288 | 49 | 819 | 674 | 352 | 53 | 1079 | 0.21 | 0.423 | 13 |
| Chang | 2008 | Lung | America | Africa American | PB | Illumina | 68 | 119 | 68 | 255 | 93 | 138 | 49 | 280 | 0.42 | 0.858 | 8 |
| Chang | 2008 | Lung | America | Latino | PB | Illumina | 60 | 44 | 9 | 113 | 138 | 127 | 34 | 299 | 0.33 | 0.561 | 7 |
| Pardini | 2008 | Colorectal | Czech | Caucasian | HB | PCR-RFLP | 334 | 177 | 21 | 532 | 356 | 153 | 23 | 532 | 0.19 | 0.211 | 11 |
| Smith | 2008 | Breast | America | African American | PB | MassARRAY | 13 | 32 | 7 | 52 | 18 | 37 | 20 | 75 | 0.51 | 0.913 | 9 |
| Hung | 2008 | Lung | World | World | Mixed | Unknown | 1852 | 1155 | 209 | 3216 | 2485 | 1510 | 286 | 4281 | 0.24 | 0.006 | 10 |
| He | 2008 | Cervical | China | Asian | HB | mismatch amplification PCR | 71 | 94 | 35 | 200 | 67 | 80 | 53 | 200 | 0.47 | 0.006 | 8 |
| Hooker | 2008 | Prostate | America | African | HB | PCR | 74 | 119 | 61 | 254 | 99 | 142 | 60 | 301 | 0.44 | 0.484 | 8 |
| Wang | 2007 | NMSC | Texas | Caucasian | HB | PCR | 146 | 89 | 11 | 246 | 200 | 119 | 10 | 329 | 0.21 | 0.121 | 8 |
| Povey | 2007 | Melanoma | Scotland | Caucasian | PB | PCR-RFLP | 314 | 169 | 24 | 507 | 252 | 162 | 27 | 441 | 0.24 | 0.887 | 13 |
| Crew | 2007 | Breast | America | Others | PB | Sequenom | 562 | 371 | 66 | 999 | 571 | 409 | 71 | 1051 | 0.26 | 0.846 | 11 |
| An | 2007 | HNC | America | Caucasian | HB | PCR | 507 | 286 | 36 | 829 | 519 | 289 | 46 | 854 | 0.22 | 0.489 | 11 |
| Jorgensen | 2007 | Breast | America | Others | PB | TaqMan | 159 | 93 | 12 | 264 | 165 | 95 | 15 | 275 | 0.23 | 0.785 | 10 |
| Mechanic | 2006 | Breast | America | African American | PB | TaqMan | 231 | 387 | 139 | 757 | 231 | 320 | 123 | 674 | 0.42 | 0.509 | 9 |
| Mechanic | 2006 | Breast | America | Caucasian | PB | TaqMan | 771 | 409 | 69 | 1249 | 661 | 412 | 60 | 1133 | 0.23 | 0.685 | 9 |
| Shen | 2006 | Breast | America | Others | PB | TaqMan | 83 | 63 | 8 | 154 | 82 | 62 | 7 | 151 | 0.25 | 0.268 | 11 |
| Sugimura | 2006 | OSCC | Japan | Asian | HB | PCR-RFLP | 43 | 59 | 20 | 122 | 77 | 112 | 52 | 241 | 0.45 | 0.348 | 5 |
| Garcia-Closas | 2006 | Bladder | Spain | Caucasian | HB | Sequencing | 629 | 434 | 78 | 1141 | 607 | 445 | 84 | 1136 | 0.27 | 0.844 | 11 |
| Li | 2006 | Melanoma | America | Caucasian | HB | PCR | 373 | 206 | 23 | 602 | 370 | 206 | 27 | 603 | 0.22 | 0.805 | 12 |
| Wu | 2006 | Bladder | America | Others | PB | TaqMan | 364 | 225 | 26 | 615 | 371 | 211 | 18 | 600 | 0.21 | 0.064 | 13 |
| Thirumaran | 2006 | BCC | Hungry, Romania, Slovakia | Caucasian | HB | TaqMan | 325 | 172 | 32 | 529 | 330 | 173 | 30 | 533 | 0.22 | 0.250 | 11 |
| Shen | 2006 | NHL | America | Others | PB | TaqMan | 260 | 170 | 34 | 464 | 352 | 169 | 29 | 550 | 0.21 | 0.146 | 13 |
| Le Morvan | 2006 | Sarcoma | France | Caucasian | HB | PCR-RFLP | 182 | 107 | 19 | 308 | 31 | 21 | 1 | 53 | 0.22 | 0.227 | 6 |
| Sakiyama | 2005 | Lung | Japan | Asian | Mixed | Pyrosequencing | 300 | 500 | 202 | 1002 | 228 | 333 | 124 | 685 | 0.42 | 0.900 | 7 |
| Shen | 2005 | Lung | China | Asian | PB | TaqMan | 38 | 52 | 26 | 116 | 38 | 46 | 25 | 109 | 0.44 | 0.133 | 10 |
| Weiss | 2005 | Endometrial | America | Caucasian | PB | PCR-RFLP | 215 | 134 | 22 | 371 | 250 | 148 | 22 | 420 | 0.23 | 0.987 | 11 |
| Blankenburg | 2005 | Melanoma | German | Caucasian | PB | PCR-RFLP | 9 | 100 | 184 | 293 | 18 | 124 | 232 | 374 | 0.79 | 0.785 | 8 |
| Sanyal | 2004 | Bladder | Sweden | Caucasian | PB | PCR-RFLP | 182 | 109 | 8 | 299 | 173 | 91 | 20 | 284 | 0.23 | 0.102 | 8 |
| Kumar | 2003 | Breast | Finland | Caucasian | PB | PCR-RFLP | 108 | 96 | 16 | 220 | 182 | 107 | 19 | 308 | 0.24 | 0.540 | 10 |
| MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium; B-NHL, B cell non-Hodgkin's lymphoma; BCL, B cell lymphoma; HNC, head and neck cancer; HCC, hepatocellular carcinoma; HD, Hodgkin’s disease; NMSC, non-melanoma skin cancer; OSCC, oral squamous cell carcinoma; BCC, basal cell carcinoma; HB, hospital based; PB, population based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; AS-PCR, allele-specific PCR. |
Meta-analysis results
As we can see in Table 2 and Figure 2, significant between-study heterogeneity was detected under all the genetic models in the overall analysis. Thus, we used random-effect model. After calculating crude odds ratios (ORs) and 95% confidence interval (CIs), we found that XPG gene rs17655 G>C polymorphism was associated with increased overall cancer susceptibility (CC vs. GG: OR=1.10, 95% CI=1.00-1.20, P=0.032; CG vs. GG: OR=1.06, 95% CI=1.02-1.11, P=0.013; CG+CC vs. GG: OR=1.07, 95% CI=1.02-1.12, P=0.004; C vs. G: OR=1.05, 95% CI=1.01-1.09, P=0.011). Stratification analysis further indicated that the XPG gene rs17655 G>C polymorphism was associated with increased risk of gastric cancer (CC vs. GG: OR=1.53, 95% CI=1.16-2.01, P=0.002; CG vs. GG: OR=1.25, 95% CI=1.02-1.53, P=0.030; CG+CC vs. GG: OR=1.32, 95% CI=1.09-1.60, P=0.005; C vs. G: OR=1.23, 95% CI=1.06-1.42, P=0.005) and colorectal cancer (CG vs. GG: OR=1.30, 95% CI=1.12-1.51, P=0.001; CG+CC vs. GG: OR=1.28, 95% CI=1.11-1.48, P=0.001; C vs. G: OR=1.16, 95% CI=1.05-1.30, P=0.011) (Supplemental Figure 1). We also checked the association in Asian (18 studies) and Caucasian (24 studies), among which ethnic groups studies were enriched. Interestingly, we only observed significant association in Asian (CC vs. GG: OR=1.25, 95% CI=1.05-1.49, P=0.013; CG vs. GG: OR=1.20, 95% CI=1.06-1.35, P=0.002; CG+CC vs. GG: OR=1.21, 95% CI=1.07-1.38, P=0.005; C vs. G: OR=1.13, 95% CI=1.03-1.23, P=0.005). Moreover, the association remained significant in the subgroups with quality score ≤ 9 (CC vs. GG: OR=1.20, 95% CI=1.04-1.39, P=0.015; CG vs. GG: OR=1.09, 95% CI=1.00-1.18, P=0.033; CG+CC vs. GG: OR=1.11, 95% CI=1.02-1.21, P=0.018; C vs. G: OR=1.07, 95% CI=1.01-1.15, P=0.065) and hospital-based studies (CC vs. GG: OR=1.19, 95% CI=1.02-1.39, P=0.028; CG vs. GG: OR=1.10, 95% CI=1.01-1.20, P=0.032; CG+CC vs. GG: OR=1.12, 95% CI=1.02-1.22, P=0.009; C vs. G: OR=1.09, 95% CI=1.02-1.16, P=0.007).
Table 2. Meta-analysis of the association between XPG gene rs17655 G>C polymorphism and overall cancer risk.
| Variables | No. of | Homozygous | | Heterozygous | | Recessive | | Dominant | | Allele |
| studies | CC vs. GG | | CG vs. GG | | CC vs. CG+GG | | CG+CC vs. GG | | C vs. G |
| | OR (95% CI) | Phet | | OR (95% CI) | Phet | | OR (95% CI) | Phet | | OR (95% CI) | Phet | | OR (95% CI) | Phet |
| All | 60 | 1.10(1.00-1.20) | 0.001 | | 1.06(1.02-1.11) | 0.040 | | 1.04(0.97-1.12) | 0.028 | | 1.07(1.02-1.12) | 0.002 | | 1.05(1.01-1.09) | 0.000 |
| Cancer type |
| Gastric | 5 | 1.53(1.16-2.01) | 0.407 | | 1.25(1.02-1.53) | 0.793 | | 1.30(0.93-1.82) | 0.131 | | 1.32(1.09-1.60) | 0.755 | | 1.23(1.06-1.42) | 0.288 |
| Breast | 11 | 1.10(0.95-1.27) | 0.613 | | 1.08(0.95-1.22) | 0.047 | | 1.04(0.92-1.19) | 0.768 | | 1.08(0.95-1.22) | 0.036 | | 1.04(0.96-1.14) | 0.073 |
| Colorectal | 4 | 1.24(0.96-1.59) | 0.395 | | 1.30(1.12-1.51) | 0.395 | | 1.06(0.84-1.34) | 0.401 | | 1.28(1.11-1.48) | 0.554 | | 1.16(1.05-1.30) | 0.875 |
| Lymphoma | 4 | 1.13(0.57-2.24) | 0.049 | | 0.98(0.69-1.41) | 0.022 | | 1.17(0.66-2.08) | 0.110 | | 0.97(0.65-1.46) | 0.004 | | 0.98(0.69-1.39) | 0.001 |
| Bladder | 6 | 0.97(0.71-1.33) | 0.177 | | 1.03(0.92-1.16) | 0.520 | | 0.93(0.70-1.24) | 0.193 | | 1.02(0.91-1.14) | 0.588 | | 1.00(0.91-1.09) | 0.636 |
| Lung | 6 | 1.26(0.92-1.73) | 0.007 | | 1.13(0.93-1.37) | 0.051 | | 1.12(0.92-1.37) | 0.136 | | 1.16(0.94-1.43) | 0.011 | | 1.11(0.96-1.28) | 0.012 |
| HNC | 3 | 0.88(0.71-1.09) | 0.819 | | 1.01(0.90-1.14) | 0.898 | | 0.90(0.74-1.10) | 0.684 | | 0.99(0.88-1.11) | 0.944 | | 0.97(0.89-1.06) | 0.984 |
| Others | 13 | 1.09(0.88-1.36) | 0.014 | | 1.04(0.95-1.14) | 0.411 | | 1.07(0.87-1.31) | 0.014 | | 1.05(0.95-1.15) | 0.226 | | 1.05(0.96-1.15) | 0.051 |
| Skin | 8 | 0.96(0.75-1.23) | 0.175 | | 0.97(0.88-1.06) | 0.793 | | 0.96(0.79-1.17) | 0.254 | | 0.96(0.88-1.05) | 0.657 | | 0.97(0.90-1.04) | 0.427 |
| Ethnicity |
| Asian | 18 | 1.25(1.05-1.49) | 0.003 | | 1.20(1.06-1.35) | 0.031 | | 1.10(0.97-1.25) | 0.044 | | 1.21(1.07-1.38) | 0.005 | | 1.13(1.03-1.23) | 0.002 |
| Caucasian | 24 | 0.98(0.87-1.10) | 0.254 | | 1.01(0.95-1.06) | 0.437 | | 0.97(0.86-1.09) | 0.230 | | 1.00(0.95-1.05) | 0.575 | | 0.99(0.95-1.04) | 0.590 |
| Quality score | |
| >9 | 25 | 0.98(0.90-1.07) | 0.872 | | 1.04(0.99-1.09) | 0.341 | | 0.97(0.90-1.05) | 0.932 | | 1.03(0.98-1.08) | 0.267 | | 1.01(0.98-1.05) | 0.447 |
| ≤9 | 35 | 1.20(1.04-1.39) | 0.000 | | 1.09(1.00-1.18) | 0.023 | | 1.10(0.98-1.24) | 0.002 | | 1.11(1.02-1.21) | 0.001 | | 1.07(1.01-1.15) | 0.000 |
| Design | |
| HB | 26 | 1.19(1.02-1.39) | 0.002 | | 1.10(1.01-1.20) | 0.031 | | 1.09(0.97-1.24) | 0.034 | | 1.12(1.02-1.22) | 0.004 | | 1.09(1.02-1.16) | 0.003 |
| PB | 31 | 1.03(0.91-1.17) | 0.079 | | 1.04(0.97-1.10) | 0.185 | | 1.00(0.90-1.12) | 0.118 | | 1.03(0.97-1.10) | 0.069 | | 1.02(0.97-1.07) | 0.022 |
| Mixed | 3 | 1.04(0.91-1.18) | 0.376 | | 1.05(0.97-1.13) | 0.690 | | 1.01(0.90-1.14) | 0.550 | | 1.04(0.97-1.12) | 0.504 | | 1.03(0.97-1.09) | 0.431 |
| HNC, Head and Neck cancer; OR, odds ratio; CI, confidence interval; Het, heterogeneity. |
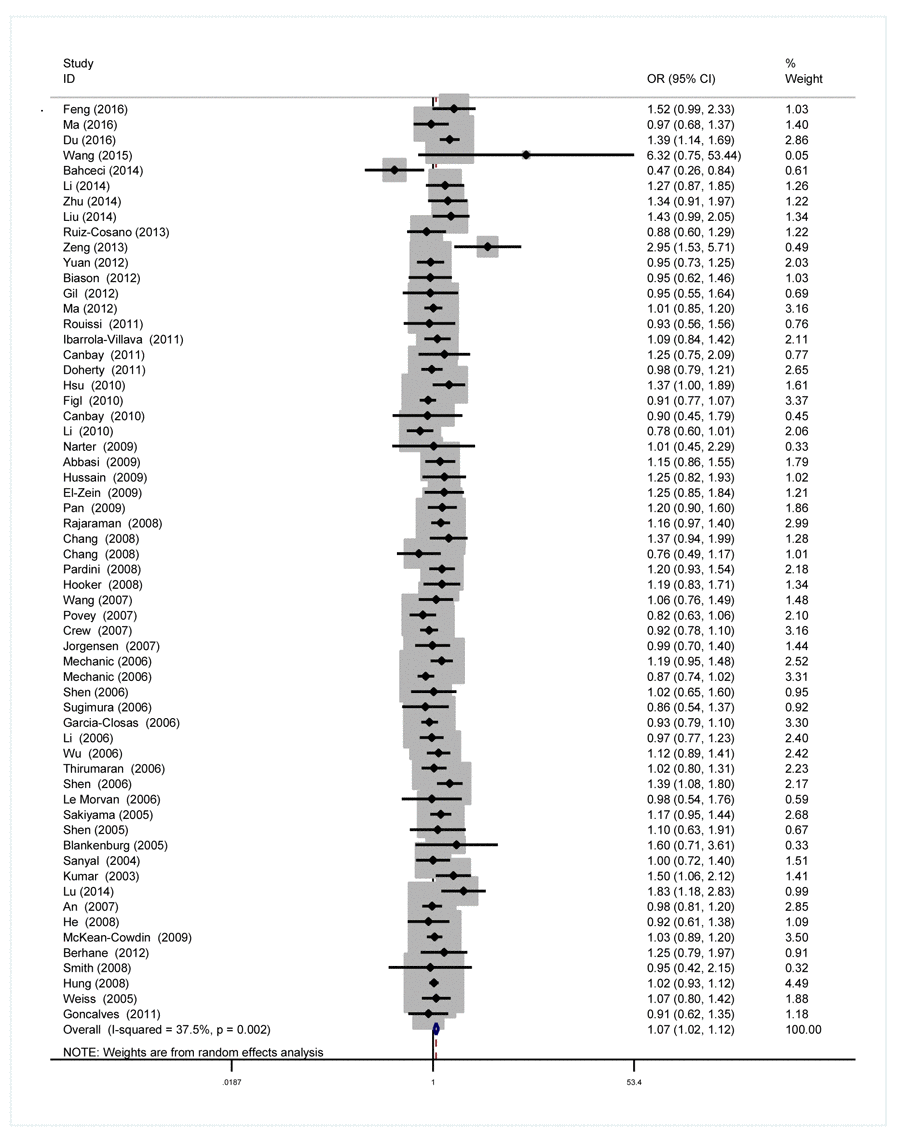
Figure 2. Forest plot for the association between the XPG rs17655 G>C polymorphism and overall cancer risk under the dominant model (CG/CC vs. GG). For each publication, the estimation of OR and its 95% CI was plotted with a box and a horizontal line. The diamonds represented the pooled ORs and 95% CIs.
Publication Bias
Symmetry in the funnel plot (Figure 3) suggested that there was no significant publication bias in this meta-analysis (CC vs. GG: P=0.808; CG vs. GG: P=0.050; CC vs. CG+GG: P=0.806; CG+CC vs. GG: P=0.047; C vs. G: P=0.240).
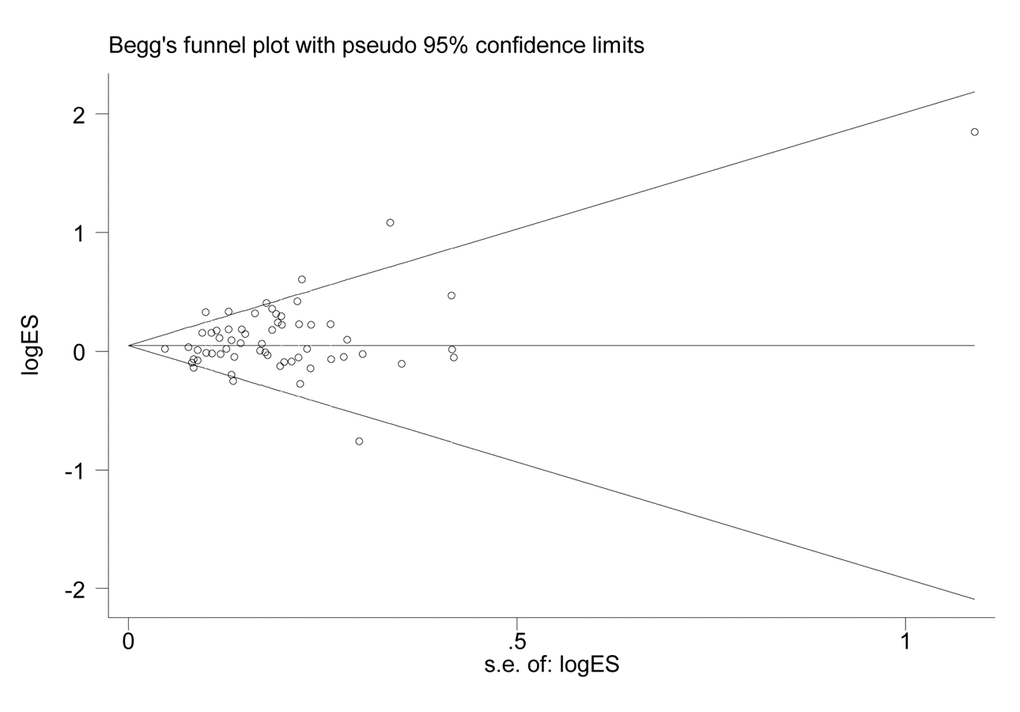
Figure 3. Funnel plot for the association between XPG gene rs17655 G>C polymorphism and overall cancer risk under the dominant model (CG/CC vs. GG).
Discussion
In the current meta-analysis, we estimated the association between the XPG gene rs17655 G>C polymorphism and cancer risk based on 60 eligible case-control studies with a total of 27,098 cancer cases and 30,535 healthy controls. Pooled risk estimates revealed that this polymorphism was significantly associated with an increased risk of overall cancer, especially with the risk of gastric cancer and colorectal cancer.
The etiology of cancer is multifactorial [1]. Abnormal accumulation of DNA mutations caused by a variety of factors might eventually trigger carcinogenic process [68]. Thus, properly repairing DNA damages in time to ensure genome stability and integrity is essential to prevent cancer. NER system includes two pathways: global genome repair and transcription-coupled repair, in both of which XPG plays a crucial role [6–8]. XPG gene, one of the eight vital genes in the NER pathway, is responsible for recognizing and excising DNA lesions on the 3’ side [3,4]. Loads of SNPs have been identified in the XPG gene over the past decades, among which the rs17655 polymorphism has revoked great attention for its association with cancer risk. The rs17655 polymorphism, leading to the replacement of aspartate with histidine at codon 1104 in ERCC5 protein, may cause an alteration in the protein function, thereby likely affecting DNA repair ability, genome integrity, and cancer predisposition.
Numerous studies were performed to explore the association between the rs17655 polymorphism and the risk of various types of cancer. Feng et al. [22] carried out a study in 2016 to investigate the roles of three SNPs (rs2094258, rs751402 and ra17655) in the XPG gene, consisting of 177 patients and 237 controls. They found that the rs17655 polymorphism was associated with an increased risk of gastric cancer. This association was reconfirmed in different types of cancer, including breast cancer by Hsu et al. [29] with 401 cases and 531controls, colorectal carcinoma by Du et al. [20] with 878 cases and 884 controls, lung cancer by Chang et al. [17] with 255 cases and 280 controls, as well as cancer of other types. However, opposite results were also frequently reported. A population-based case-control study containing 196 gastric cases and 397 controls subjects conducted by Hussain et al. [31] revealed that the XPG rs17655 polymorphism might be associated with reduced gastric cancer risk. Additionally, Ruiz-Cosano et al. [68] reported that this polymorphism did not seem to play a major role in lymphoma susceptibility after studying 213 cases and 214 controls. Ma et al. [62] selected 320 cases and 294 controls and found that the rs17655 polymorphism might not confer susceptibility to breast cancer after adjusting for potential confounding factors. Several meta-analyses were also conducted, and unfortunately the results were still inconsistent [88–91]. As contradictory results were produced, we performed this meta-analysis to draw a more precise conclusion by including larger sample size and different cancer types from 60 studies. Our result indicated that this polymorphism may increase the risk of overall cancer, especially the risk of gastric cancer and colorectal cancer. The biological function of the rs17655 remains obscure. This polymorphism has been intensively studied for its association with cancer risk as a tagger. It was predicated to be a harmful variant by a sequence homology-based tool [92]. Moreover, its functional potential was further confirmed by SIFT algorithms (scale invariant feature transform) and SNPs3D tools (http://compbio.cs.queensu.ca/F-SNP/) [93]; however, solid in vitro and in vivo data are needed to elucidate biological function of this variant.
There are advantages that strengthened the robustness of our findings. First, we searched five databases to include most of the publications written in English or Chinese. The large sample size provided adequate statistical power. Second, stratified analyses were performed by cancer type, quality score, and source of control. Third, we used the Begg’s funnel plot and Egger’s linear regression test to assess the possible publication bias.
However, several limitations still existed in this meta-analysis. Firstly, selection bias might occur because only publications written in English or Chinese were included. Researches in other languages were missed. Secondly, the number of individual studies for some cancer types, like HNC and prostate cancer (<5 studies), may be inadequate. Third, more than half of included studies had relative low quality scores (≤ 9). Our results should be interpreted cautiously. Further studies with high quality scores are needed to verify the real association.
Additionally, age, sex, living habits, virus infections or some environmental factors may also influence cancer risk. Our findings based on unadjusted estimates for lack of access to original data might suffer from potential confounding bias. Therefore, the results should be interpreted with caution. Finally, lack of biological evidence of the implication of the rs17655 polymorphism in cancer is also a drawback of the study. Mechanistic studies of the rs17655 polymorphism with cancer should be performed in the future.
In conclusion, this meta-analysis suggests that the XPG rs17655 G>C polymorphism is significantly associated with an increased overall cancer risk, especially with the risk of gastric cancer and colorectal cancer. Moreover, large-scale, well-designed studies in different cancers should be conducted to corroborate our findings.
Materials and Methods
Publication search
We searched for relevant articles using the following terms: “ERCC5 or XPG”, “polymorphism or variant”, and “cancer or carcinoma or neoplasm or malignance” in PubMed, EMBASE, CNKI, WANFANG, and Vip databases (the last search was performed on June 17, 2016). We also manually searched the references of the retrieved publications for additional relevant eligible studies.
Inclusion and Exclusion criteria
The publications contained in the meta-analysis had to meet the following criteria: (1) the study was only written in English or Chinese; (2) the study investigated the association between the XPG gene rs17655 polymorphism and the risk of one or more types of cancer; (3) case-control study. If studies had overlapping subjects, the publication including the largest number of individuals were selected.
Exclusion criteria were as follows (1) the study did not report sufficient genotype data to calculate odds ratio (OR) and 95% confidence interval (CI); (2) the study included survival data only. (3) the genotype frequencies of the rs17655 G>C and other polymorphisms were deviated from Hardy-Weinberg equilibrium (HWE) in the controls.
Data Extraction and quality assessment
Two investigators (Chen SS and Zhao J) extracted the following information from each publication independently: first author, publication year, cancer type, country of origin, race, genotyping method, source of controls (hospital-based, population-based and mixed), the genotype counts of cases and controls for the rs17655 G>C polymorphism. We also calculated the score of each publication based on the quality score assessment as described before [94]. All contradictory information was discussed when necessary.
Statistical analysis
We evaluated crude ORs and 95% CIs to assess the association between XPG rs17655 G>C polymorphism and overall cancer risk under the homozygous (CC vs. GG), heterozygous (CG vs. GG), recessive (CC vs. CG+GG), dominant (CG+CC vs. GG), and allele contrast (C vs. G) models. We carried out stratification analyses by cancer type (if one cancer type were investigated in less than three studies, we termed this type as “others”), score (>9 and ≤9), and study design (if a study contained both hospital-based controls and population-based subjects, we termed the study design as “mixed”). We also calculated between-study heterogeneity using the Chi square-based Q-test. When P>0.1 indicating lack of heterogeneity, a fixed-effect model was adopted. Otherwise, a random-effect model would be applied [94]. The potential publication bias was evaluated by Begg’s funnel plot [95] and Egger’s linear regression test [96]. All of the P values were two-tailed. P<0.05 was considered statistically significant. All data analyses were performed by the STATA software (Version 12.0; Stata Corporation, College Station, TX).
Acknowledgements
This study was supported by grants from the Scientific Research Foundation of Wenzhou (2015Y0492), Zhejiang Provincial Medical and Health Science and Technology plan (2009A148), Zhejiang Provincial Science and Technology Animal Experimental Platform Project (016C37113), Scientific Research Fund of Wenling Science and Technology Bureau (2015C31BA0049), and Natural Science Foundation of Heilongjiang Province (H2015049).
Conflicts of Interest
We had no conflicts of interest to declare.
References
-
1.
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108. https://doi.org/10.3322/caac.21262 [PubMed]
-
2.
Le Morvan V, Longy M, Bonaïti-Pellié C, Bui B, Houédé N, Coindre JM, Robert J, Pourquier P. Genetic polymorphisms of the XPG and XPD nucleotide excision repair genes in sarcoma patients. Int J Cancer. 2006; 119:1732–35. https://doi.org/10.1002/ijc.22009 [PubMed]
-
3.
Bernstein C, Bernstein H, Payne CM, Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002; 511:145–78. https://doi.org/10.1016/S1383-5742(02)00009-1 [PubMed]
-
4.
Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005; 577:275–83. https://doi.org/10.1016/j.mrfmmm.2005.03.007 [PubMed]
-
5.
Friedberg EC, Bond JP, Burns DK, Cheo DL, Greenblatt MS, Meira LB, Nahari D, Reis AM. Defective nucleotide excision repair in xpc mutant mice and its association with cancer predisposition. Mutat Res. 2000; 459:99–108. https://doi.org/10.1016/S0921-8777(99)00068-3 [PubMed]
-
6.
Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst). 2002; 1:95–107. https://doi.org/10.1016/S1568-7864(01)00008-8 [PubMed]
-
7.
O’Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994; 371:432–35. https://doi.org/10.1038/371432a0 [PubMed]
-
8.
Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sci. 2007; 4:59–71. https://doi.org/10.7150/ijms.4.59 [PubMed]
-
9.
Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, Popanda O. Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer. 2009; 125:1431–39. https://doi.org/10.1002/ijc.24442 [PubMed]
-
10.
An J, Liu Z, Hu Z, Li G, Wang LE, Sturgis EM, El-Naggar AK, Spitz MR, Wei Q. Potentially functional single nucleotide polymorphisms in the core nucleotide excision repair genes and risk of squamous cell carcinoma of the head and neck. Cancer Epidemiol Biomarkers Prev. 2007; 16:1633–38. https://doi.org/10.1158/1055-9965.EPI-07-0252 [PubMed]
-
11.
Bahceci A, Paydas S, Tanriverdi K, Ergin M, Seydaoglu G, Ucar G. DNA repair gene polymorphisms in B cell non-Hodgkin’s lymphoma. Tumour Biol. 2015; 36:2155–61. https://doi.org/10.1007/s13277-014-2825-9 [PubMed]
-
12.
Berhane N, Sobti RC, Mahdi SA. DNA repair genes polymorphism (XPG and XRCC1) and association of prostate cancer in a north Indian population. Mol Biol Rep. 2012; 39:2471–79. https://doi.org/10.1007/s11033-011-0998-5 [PubMed]
-
13.
Biason P, Hattinger CM, Innocenti F, Talamini R, Alberghini M, Scotlandi K, Zanusso C, Serra M, Toffoli G. Nucleotide excision repair gene variants and association with survival in osteosarcoma patients treated with neoadjuvant chemotherapy. Pharmacogenomics J. 2012; 12:476–83. https://doi.org/10.1038/tpj.2011.33 [PubMed]
-
14.
Blankenburg S, König IR, Moessner R, Laspe P, Thoms KM, Krueger U, Khan SG, Westphal G, Volkenandt M, Neumann C, Ziegler A, Kraemer KH, Reich K, Emmert S. No association between three xeroderma pigmentosum group C and one group G gene polymorphisms and risk of cutaneous melanoma. Eur J Hum Genet. 2005; 13:253–55. https://doi.org/10.1038/sj.ejhg.5201296 [PubMed]
-
15.
Canbay E, Agachan B, Gulluoglu M, Isbir T, Balik E, Yamaner S, Bulut T, Cacina C, Eraltan IY, Yilmaz A, Bugra D. Possible associations of APE1 polymorphism with susceptibility and HOGG1 polymorphism with prognosis in gastric cancer. Anticancer Res. 2010; 30:1359–64. [PubMed]
-
16.
Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, Gulluoglu M, Balik E, Bulut T, Yamaner S, Bugra D. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011; 27:1295–302. https://doi.org/10.1185/03007995.2011.573544 [PubMed]
-
17.
Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CPJr, Seldin MF, Kelsey KT, Kittles RA, Silva G, Wiencke JK. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer. 2008; 123:2095–104. https://doi.org/10.1002/ijc.23801 [PubMed]
-
18.
Crew KD, Gammon MD, Terry MB, Zhang FF, Zablotska LB, Agrawal M, Shen J, Long CM, Eng SM, Sagiv SK, Teitelbaum SL, Neugut AI, Santella RM. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007; 16:2033–41. https://doi.org/10.1158/1055-9965.EPI-07-0096 [PubMed]
-
19.
Doherty JA, Weiss NS, Fish S, Fan W, Loomis MM, Sakoda LC, Rossing MA, Zhao LP, Chen C. Polymorphisms in nucleotide excision repair genes and endometrial cancer risk. Cancer Epidemiol Biomarkers Prev. 2011; 20:1873–82. https://doi.org/10.1158/1055-9965.EPI-11-0119 [PubMed]
-
20.
Du HN, Zhu LJ, Du ML, Wang ML, Shu YQ. The association between XPG Asp1104His polymorphism and colorectal cancer risk in the Chinese population. Chin J Colorec Dis. 2016; 5:40–46. Electronic Edition. https://doi.org/10.3877/cma.j.issn.2095-3224.2016.01.08
-
21.
El-Zein R, Monroy CM, Etzel CJ, Cortes AC, Xing Y, Collier AL, Strom SS. Genetic polymorphisms in DNA repair genes as modulators of Hodgkin disease risk. Cancer. 2009; 115:1651–59. https://doi.org/10.1002/cncr.24205 [PubMed]
-
22.
Feng YB, Fan DQ, Yu J, Bie YK. Association between XPG gene polymorphisms and development of gastric cancer risk in a Chinese population. Genet Mol Res. 2016; 15. https://doi.org/10.4238/gmr.15027877 [PubMed]
-
23.
Figl A, Scherer D, Nagore E, Bermejo JL, Botella-Estrada R, Gast A, Thirumaran RK, Planelles D, Hemminki K, Schadendorf D, Kumar R. Single-nucleotide polymorphisms in DNA-repair genes and cutaneous melanoma. Mutat Res. 2010; 702:8–16. https://doi.org/10.1016/j.mrgentox.2010.06.011 [PubMed]
-
24.
García-Closas M, Malats N, Real FX, Welch R, Kogevinas M, Chatterjee N, Pfeiffer R, Silverman D, Dosemeci M, Tardón A, Serra C, Carrato A, García-Closas R, et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2006; 15:536–42. https://doi.org/10.1158/1055-9965.EPI-05-0749 [PubMed]
-
25.
Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, Laczmanska I, Leszczynski P, Grzebieniak Z, Sasiadek MM. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual’s susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012; 39:527–34. https://doi.org/10.1007/s11033-011-0767-5 [PubMed]
-
26.
Gonçalves FT, Francisco G, de Souza SP, Luiz OC, Festa-Neto C, Sanches JA, Chammas R, Gattas GJ, Eluf-Neto J. European ancestry and polymorphisms in DNA repair genes modify the risk of melanoma: a case-control study in a high UV index region in Brazil. J Dermatol Sci. 2011; 64:59–66. https://doi.org/10.1016/j.jdermsci.2011.06.003 [PubMed]
-
27.
He X, Ye F, Zhang J, Cheng Q, Shen J, Chen H. Susceptibility of XRCC3, XPD, and XPG genetic variants to cervical carcinoma. Pathobiology. 2008; 75:356–63. https://doi.org/10.1159/000164220 [PubMed]
-
28.
Hooker S, Bonilla C, Akereyeni F, Ahaghotu C, Kittles RA. NAT2 and NER genetic variants and sporadic prostate cancer susceptibility in African Americans. Prostate Cancer Prostatic Dis. 2008; 11:349–56. https://doi.org/10.1038/sj.pcan.4501027 [PubMed]
-
29.
Ming-Shiean H, Yu JC, Wang HW, Chen ST, Hsiung CN, Ding SL, Wu PE, Shen CY, Cheng CW. Synergistic effects of polymorphisms in DNA repair genes and endogenous estrogen exposure on female breast cancer risk. Ann Surg Oncol. 2010; 17:760–71. https://doi.org/10.1245/s10434-009-0802-0 [PubMed]
-
30.
Hung RJ, Christiani DC, Risch A, Popanda O, Haugen A, Zienolddiny S, Benhamou S, Bouchardy C, Lan Q, Spitz MR, Wichmann HE, LeMarchand L, Vineis P, et al, and International Lung Cancer Consortium. International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev. 2008; 17:3081–89. https://doi.org/10.1158/1055-9965.EPI-08-0411 [PubMed]
-
31.
Hussain SK, Mu LN, Cai L, Chang SC, Park SL, Oh SS, Wang Y, Goldstein BY, Ding BG, Jiang Q, Rao J, You NC, Yu SZ, et al. Genetic variation in immune regulation and DNA repair pathways and stomach cancer in China. Cancer Epidemiol Biomarkers Prev. 2009; 18:2304–09. https://doi.org/10.1158/1055-9965.EPI-09-0233 [PubMed]
-
32.
Ibarrola-Villava M, Peña-Chilet M, Fernandez LP, Aviles JA, Mayor M, Martin-Gonzalez M, Gomez-Fernandez C, Casado B, Lazaro P, Lluch A, Benitez J, Lozoya R, Boldo E, et al. Genetic polymorphisms in DNA repair and oxidative stress pathways associated with malignant melanoma susceptibility. Eur J Cancer. 2011; 47:2618–25. https://doi.org/10.1016/j.ejca.2011.05.011 [PubMed]
-
33.
Jorgensen TJ, Visvanathan K, Ruczinski I, Thuita L, Hoffman S, Helzlsouer KJ. Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat. 2007; 101:65–71. https://doi.org/10.1007/s10549-006-9263-3 [PubMed]
-
34.
Kumar R, Höglund L, Zhao C, Försti A, Snellman E, Hemminki K. Single nucleotide polymorphisms in the XPG gene: determination of role in DNA repair and breast cancer risk. Int J Cancer. 2003; 103:671–75. https://doi.org/10.1002/ijc.10870 [PubMed]
-
35.
Li C, Hu Z, Liu Z, Wang LE, Strom SS, Gershenwald JE, Lee JE, Ross MI, Mansfield PF, Cormier JN, Prieto VG, Duvic M, Grimm EA, Wei Q. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2006; 15:2526–32. https://doi.org/10.1158/1055-9965.EPI-06-0672 [PubMed]
-
36.
Li LM, Zeng XY, Ji L, Fan XJ, Li YQ, Hu XH, Qiu XQ, Yu HP. [Association of XPC and XPG polymorphisms with the risk of hepatocellular carcinoma]. Zhonghua Gan Zang Bing Za Zhi. 2010; 18:271–75. https://doi.org/10.3760/cma.j.issn.1007-3418.2010.04.009 [PubMed]
-
37.
Li XC, Xiong JG, Cheng ZW, Wu J, Liu QS. Association between XPG polymorphisms and risk of gastric cancer. Chin J Gastroenterol Hepatol. 2014; 23:259–62.
-
38.
Liu J. The polymorphism of XPG His1104Asp gene and its risk of gastric cancer. Mod Prev Med. 2014; 41:2895–98.
-
39.
Rajaraman P, Hutchinson A, Wichner S, Black PM, Fine HA, Loeffler JS, Selker RG, Shapiro WR, Rothman N, Linet MS, Inskip PD. DNA repair gene polymorphisms and risk of adult meningioma, glioma, and acoustic neuroma. Neuro-oncol. 2010; 12:37–48. https://doi.org/10.1093/neuonc/nop012 [PubMed]
-
40.
Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Chen C. Nucleotide excision repair genotype and the incidence of endometrial cancer: effect of other risk factors on the association. Gynecol Oncol. 2006; 103:891–96. https://doi.org/10.1016/j.ygyno.2006.05.020 [PubMed]
-
41.
Chen M, Kamat AM, Huang M, Grossman HB, Dinney CP, Lerner SP, Wu X, Gu J. High-order interactions among genetic polymorphisms in nucleotide excision repair pathway genes and smoking in modulating bladder cancer risk. Carcinogenesis. 2007; 28:2160–65. https://doi.org/10.1093/carcin/bgm167 [PubMed]
-
42.
Cui Y, Morgenstern H, Greenland S, Tashkin DP, Mao J, Cao W, Cozen W, Mack TM, Zhang ZF. Polymorphism of Xeroderma Pigmentosum group G and the risk of lung cancer and squamous cell carcinomas of the oropharynx, larynx and esophagus. Int J Cancer. 2006; 118:714–20. https://doi.org/10.1002/ijc.21413 [PubMed]
-
43.
Hill DA, Wang SS, Cerhan JR, Davis S, Cozen W, Severson RK, Hartge P, Wacholder S, Yeager M, Chanock SJ, Rothman N. Risk of non-Hodgkin lymphoma (NHL) in relation to germline variation in DNA repair and related genes. Blood. 2006; 108:3161–67. https://doi.org/10.1182/blood-2005-01-026690 [PubMed]
-
44.
Hung RJ, Baragatti M, Thomas D, McKay J, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, Bencko V, et al. Inherited predisposition of lung cancer: a hierarchical modeling approach to DNA repair and cell cycle control pathways. Cancer Epidemiol Biomarkers Prev. 2007; 16:2736–44. https://doi.org/10.1158/1055-9965.EPI-07-0494 [PubMed]
-
45.
Joshi AD, Corral R, Siegmund KD, Haile RW, Le Marchand L, Martínez ME, Ahnen DJ, Sandler RS, Lance P, Stern MC. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009; 30:472–79. https://doi.org/10.1093/carcin/bgn260 [PubMed]
-
46.
Liu Y, Scheurer ME, El-Zein R, Cao Y, Do KA, Gilbert M, Aldape KD, Wei Q, Etzel C, Bondy ML. Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol Biomarkers Prev. 2009; 18:204–14. https://doi.org/10.1158/1055-9965.EPI-08-0632 [PubMed]
-
47.
Mort R, Mo L, McEwan C, Melton DW. Lack of involvement of nucleotide excision repair gene polymorphisms in colorectal cancer. Br J Cancer. 2003; 89:333–37. https://doi.org/10.1038/sj.bjc.6601061 [PubMed]
-
48.
Shen M, Purdue MP, Kricker A, Lan Q, Grulich AE, Vajdic CM, Turner J, Whitby D, Chanock S, Rothman N, Armstrong BK. Polymorphisms in DNA repair genes and risk of non-Hodgkin’s lymphoma in New South Wales, Australia. Haematologica. 2007; 92:1180–85. https://doi.org/10.3324/haematol.11324 [PubMed]
-
49.
Wang M, Chu H, Zhang Z, Wei Q. Molecular epidemiology of DNA repair gene polymorphisms and head and neck cancer. J Biomed Res. 2013; 27:179–92. https://doi.org/10.7555/JBR.27.20130034 [PubMed]
-
50.
Wen H, Ding Q, Fang ZJ, Xia GW, Fang J. [Correlation of DNA repair genetic polymorphisms and non muscle-invasive bladder cancer risk]. Chinese Journal of Urology. 2009; 30:336–39.
-
51.
Wen H, Ding Q, Fang ZJ, Xia GW, Fang J. Population study of genetic polymorphisms and superficial bladder cancer risk in Han-Chinese smokers in Shanghai. Int Urol Nephrol. 2009; 41:855–64. https://doi.org/10.1007/s11255-009-9560-y [PubMed]
-
52.
Guo NN, Du HN, Zhu LJ. [The relation between XPG G3507C polymorphism and the sensitivity toplatinum-based chemotherapy in patients with colon cancer with III stage]. Journal of Modern Oncology. 2014; 22:2142–44.
-
53.
Xie Wm, Zai Y, Zhou Gq, Zhang Hx, Huang Wf, Liao Xl, Wang Hx, Qin Fh, Liang Ar, Xie Ya, Cui Y. [Association between the DNA repair gene XPG His1104Asp polymorphism and clinical phenotypes and outcome of hepatocellular carcinoma]. China Oncology. 2012; 22:458–63.
-
54.
Yu QZ, Han JX, Pan JH, Sheng LJ, Wu JM, Huang HN. [Correlation between XPG,MDR-1 gene mononucleotide polymorphisms and responsiveness of advanced NSCLC to platinum-based chemotherapy]. Shandong Yiyao. 2007; 47:7–9.
-
55.
Zhang W, Wu Fx, Wang Q, Li Dj, Li L. [Analysis of genetic polymorphisms in nucleotide excision repair system in platinum drug resistant and non-resistant ovarian cancer cells]. Sichuan Journal of Cancer Control. 2011; 24:284–89.
-
56.
Zhang W, Wu Fx, Wang Q, Li Dj, Li L. [The relation between XPG genetic polymorphism and the sensitivity toplatinum-based chemotherapy in patients with ovarian cancer]. Zhongguo Shiyong Fuke Yu Chanke Zazhi. 2012; 28:369–71.
-
57.
Guo BW, Yang L, Zhao R, Hao SZ. Association between ERCC5 gene polymorphisms and gastric cancer risk. Genet Mol Res. 2016; 15. https://doi.org/10.4238/gmr.15027828 [PubMed]
-
58.
Jeon HS, Kim KM, Park SH, Lee SY, Choi JE, Lee GY, Kam S, Park RW, Kim IS, Kim CH, Jung TH, Park JY. Relationship between XPG codon 1104 polymorphism and risk of primary lung cancer. Carcinogenesis. 2003; 24:1677–81. https://doi.org/10.1093/carcin/bgg120 [PubMed]
-
59.
Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN, Liu-Mares W, Hu JJ. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008; 29:2132–38. https://doi.org/10.1093/carcin/bgn193 [PubMed]
-
60.
Sun K, Gong A, Liang P. Predictive impact of genetic polymorphisms in DNA repair genes on susceptibility and therapeutic outcomes to colorectal cancer patients. Tumour Biol. 2015; 36:1549–59. https://doi.org/10.1007/s13277-014-2721-3 [PubMed]
-
61.
Wen SX, Tang PZ, Zhang XM, Zhao D, Guo YL, Tan W, Lin DX. [Association between genetic polymorphism in xeroderma pigmentosum G gene and risks of laryngeal and hypopharyngeal carcinomas]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2006; 28:703–06. [PubMed]
-
62.
Ma SH, Ling FH, Sun YX, Chen SF, Li Z. Investigation on the role of XPG gene polymorphisms in breast cancer risk in a Chinese population. Genet Mol Res. 2016; 15. https://doi.org/10.4238/gmr.15028066 [PubMed]
-
63.
Mechanic LE, Millikan RC, Player J, de Cotret AR, Winkel S, Worley K, Heard K, Heard K, Tse CK, Keku T. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis. 2006; 27:1377–85. https://doi.org/10.1093/carcin/bgi330 [PubMed]
-
64.
Rajaraman P, Bhatti P, Doody MM, Simon SL, Weinstock RM, Linet MS, Rosenstein M, Stovall M, Alexander BH, Preston DL, Sigurdson AJ. Nucleotide excision repair polymorphisms may modify ionizing radiation-related breast cancer risk in US radiologic technologists. Int J Cancer. 2008; 123:2713–16. https://doi.org/10.1002/ijc.23779 [PubMed]
-
65.
Shen J, Desai M, Agrawal M, Kennedy DO, Senie RT, Santella RM, Terry MB. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev. 2006; 15:1614–19. https://doi.org/10.1158/1055-9965.EPI-06-0218 [PubMed]
-
66.
Wang H, Wang T, Guo H, Zhu G, Yang S, Hu Q, Du Y, Bai X, Chen X, Su H. Association analysis of ERCC5 gene polymorphisms with risk of breast cancer in Han women of northwest China. Breast Cancer. 2016; 23:479–85. https://doi.org/10.1007/s12282-015-0590-2 [PubMed]
-
67.
Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, Hanova M, Slyskova J, Tulupova E, Kumar R, Bortlik M, Barale R, Hemminki K, Vodicka P. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008; 638:146–53. https://doi.org/10.1016/j.mrfmmm.2007.09.008 [PubMed]
-
68.
Ruiz-Cosano J, Torres-Moreno D, Conesa-Zamora P. Influence of polymorphisms in ERCC5, XPA and MTR DNA repair and synthesis genes in B-cell lymphoma risk. A case-control study in Spanish population. J BUON. 2013; 18:486–90. [PubMed]
-
69.
Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, Wang SS, Holford TR, Leaderer B, Yeager M, Welch R, Kang D, Boyle P, Zhang B, et al. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Hum Genet. 2006; 119:659–68. https://doi.org/10.1007/s00439-006-0177-2 [PubMed]
-
70.
Narter KF, Ergen A, Agaçhan B, Görmüs U, Timirci O, Isbir T. Bladder cancer and polymorphisms of DNA repair genes (XRCC1, XRCC3, XPD, XPG, APE1, hOGG1). Anticancer Res. 2009; 29:1389–93. [PubMed]
-
71.
Rouissi K, Ouerhani S, Hamrita B, Bougatef K, Marrakchi R, Cherif M, Ben Slama MR, Bouzouita M, Chebil M, Ben Ammar Elgaaied A. Smoking and polymorphisms in xenobiotic metabolism and DNA repair genes are additive risk factors affecting bladder cancer in Northern Tunisia. Pathol Oncol Res. 2011; 17:879–86. https://doi.org/10.1007/s12253-011-9398-3 [PubMed]
-
72.
Sanyal S, Festa F, Sakano S, Zhang Z, Steineck G, Norming U, Wijkström H, Larsson P, Kumar R, Hemminki K. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004; 25:729–34. https://doi.org/10.1093/carcin/bgh058 [PubMed]
-
73.
Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, Zhang Q, Millikan RE, Lerner S, Dinney CP, Spitz MR. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006; 78:464–79. https://doi.org/10.1086/500848 [PubMed]
-
74.
Zhu CB, Deng QW, Xu Z, Zhu JG. A study on the association of polymorphisms in XPC, XPD, XPG risk of bladder cancer and its clinicopathological parameters. Acta Universitatis Medicinalis Nanjing. 2014; 34:1355–59.
-
75.
Sakiyama T, Kohno T, Mimaki S, Ohta T, Yanagitani N, Sobue T, Kunitoh H, Saito R, Shimizu K, Hirama C, Kimura J, Maeno G, Hirose H, et al. Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer. 2005; 114:730–37. https://doi.org/10.1002/ijc.20790 [PubMed]
-
76.
Shen M, Berndt SI, Rothman N, Demarini DM, Mumford JL, He X, Bonner MR, Tian L, Yeager M, Welch R, Chanock S, Zheng T, Caporaso N, Lan Q. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer. 2005; 116:768–73. https://doi.org/10.1002/ijc.21117 [PubMed]
-
77.
Zeng H, Kang MF. Association of XPA A23G and His1104Asp polymorphisms with lung cancer susceptibility. Guangdong Yixue. 2013; 34:413–16.
-
78.
Povey JE, Darakhshan F, Robertson K, Bisset Y, Mekky M, Rees J, Doherty V, Kavanagh G, Anderson N, Campbell H, MacKie RM, Melton DW. DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis. 2007; 28:1087–93. https://doi.org/10.1093/carcin/bgl257 [PubMed]
-
79.
Thirumaran RK, Bermejo JL, Rudnai P, Gurzau E, Koppova K, Goessler W, Vahter M, Leonardi GS, Clemens F, Fletcher T, Hemminki K, Kumar R. Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis. 2006; 27:1676–81. https://doi.org/10.1093/carcin/bgi381 [PubMed]
-
80.
Wang LE, Li C, Strom SS, Goldberg LH, Brewster A, Guo Z, Qiao Y, Clayman GL, Lee JJ, El-Naggar AK, Prieto VG, Duvic M, Lippman SM, et al. Repair capacity for UV light induced DNA damage associated with risk of nonmelanoma skin cancer and tumor progression. Clin Cancer Res. 2007; 13:6532–39. https://doi.org/10.1158/1078-0432.CCR-07-0969 [PubMed]
-
81.
Ma H, Yu H, Liu Z, Wang LE, Sturgis EM, Wei Q. Polymorphisms of XPG/ERCC5 and risk of squamous cell carcinoma of the head and neck. Pharmacogenet Genomics. 2012; 22:50–57. https://doi.org/10.1097/FPC.0b013e32834e3cf6 [PubMed]
-
82.
Yuan H. Genetic Susceptibility of Head and Neck Cancer in Chinese Han Population. Nanjing Medical University. 2012.
-
83.
Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Voigt LF, Chen C. Interindividual variation in nucleotide excision repair genes and risk of endometrial cancer. Cancer Epidemiol Biomarkers Prev. 2005; 14:2524–30. https://doi.org/10.1158/1055-9965.EPI-05-0414 [PubMed]
-
84.
Lu B, Li J, Gao Q, Yu W, Yang Q, Li X. Laryngeal cancer risk and common single nucleotide polymorphisms in nucleotide excision repair pathway genes ERCC1, ERCC2, ERCC3, ERCC4, ERCC5 and XPA. Gene. 2014; 542:64–68. https://doi.org/10.1016/j.gene.2014.02.043 [PubMed]
-
85.
Pan J, Lin J, Izzo JG, Liu Y, Xing J, Huang M, Ajani JA, Wu X. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis. 2009; 30:785–92. https://doi.org/10.1093/carcin/bgp058 [PubMed]
-
86.
Sugimura T, Kumimoto H, Tohnai I, Fukui T, Matsuo K, Tsurusako S, Mitsudo K, Ueda M, Tajima K, Ishizaki K. Gene-environment interaction involved in oral carcinogenesis: molecular epidemiological study for metabolic and DNA repair gene polymorphisms. J Oral Pathol Med. 2006; 35:11–18. https://doi.org/10.1111/j.1600-0714.2005.00364.x [PubMed]
-
87.
McKean-Cowdin R, Barnholtz-Sloan J, Inskip PD, Ruder AM, Butler M, Rajaraman P, Razavi P, Patoka J, Wiencke JK, Bondy ML, Wrensch M. Associations between polymorphisms in DNA repair genes and glioblastoma. Cancer Epidemiol Biomarkers Prev. 2009; 18:1118–26. https://doi.org/10.1158/1055-9965.EPI-08-1078 [PubMed]
-
88.
Aggarwal N, Donald ND, Malik S, Selvendran SS, McPhail MJ, Monahan KJ. The association of low-penetrance variants in DNA repair genes with colorectal cancer: a systematic review and meta-analysis. Clin Transl Gastroenterol. 2017; 8:e109. https://doi.org/10.1038/ctg.2017.35 [PubMed]
-
89.
Liang Y, Deng J, Xiong Y, Wang S, Xiong W. Genetic association between ERCC5 rs17655 polymorphism and lung cancer risk: evidence based on a meta-analysis. Tumour Biol. 2014; 35:5613–18. https://doi.org/10.1007/s13277-014-1742-2 [PubMed]
-
90.
Qian T, Zhang B, Qian C, He Y, Li Y. Association between common polymorphisms in ERCC gene and glioma risk: a meta-analysis of 15 studies. Medicine (Baltimore). 2017; 96:e6832. https://doi.org/10.1097/MD.0000000000006832 [PubMed]
-
91.
Xu Y, Jiao G, Wei L, Wang N, Xue Y, Lan J, Wang Y, Liu C, Lou M. Current evidences on the XPG Asp1104His polymorphism and melanoma susceptibility: a meta-analysis based on case-control studies. Mol Genet Genomics. 2015; 290:273–79. https://doi.org/10.1007/s00438-014-0917-2 [PubMed]
-
92.
Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001; 11:863–74. https://doi.org/10.1101/gr.176601 [PubMed]
-
93.
Zhu ML, Wang M, Cao ZG, He J, Shi TY, Xia KQ, Qiu LX, Wei QY. Association between the ERCC5 Asp1104His polymorphism and cancer risk: a meta-analysis. PLoS One. 2012; 7:e36293. https://doi.org/10.1371/journal.pone.0036293 [PubMed]
-
94.
He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, Chen W, Jia WH. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014; 4:6159. https://doi.org/10.1038/srep06159 [PubMed]
-
95.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994; 50:1088–101. https://doi.org/10.2307/2533446 [PubMed]
-
96.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997; 315:629–34. https://doi.org/10.1136/bmj.315.7109.629 [PubMed]