Effect of low and high doses of radiation on cells: analysis of differentially expressed genes and pathways
Gene expression samples from replicatively aged fibroblasts [11] and those taken from different patients [12] were stratified into groups by the number of passages and donor’s age, respectively (Table 1). Samples of fibroblasts that were irradiated with X-ray [9] were stratified into functional groups by the dose and time after exposure (Table 1). For each of the functional groups we discovered significantly differentially expressed genes and signaling pathways, identified by the iPanda algorithm [12] and listed in the Table 1.
Table 1. Analysis of differentially expressed genes and metabolic pathways for the selected datasets.
| Study | Age groups | Irradiation dose, time after exposure | Differentially expressed genes,
p-value < 0.01 | Signaling pathways,
p-value < 0.05 |
| EMTAB2086_70_vs_30 | 70/30 cycles | - | 1676 | 387 |
| EMTAB2086_80_vs_30 | 80/30 cycles | - | 4217 | 1328 |
| GSE55118_middle_vs_young | 30-50/<30 years | - | 603 | 29 |
| GSE55118_old_vs_young | >50/<30 years | - | 442 | 24 |
| GSE59861_12h_2Gy | - | 12 h., 2 Gy | 485 | 24 |
| GSE59861_12h_5cGy | - | 12 h., 5 cGy | 359 | 11 |
| GSE59861_24h_2Gy | - | 24 h., 2 Gy | 1201 | 108 |
| GSE59861_24h_5cGy | - | 24 h., 5 cGy | 905 | 41 |
| GSE59861_3h_2Gy | - | 3 h., 2 Gy | 555 | 44 |
| GSE59861_3h_5cGy | - | 3 h., 5 cGy | 70 | 0 |
| GSE59861_6h_2Gy | - | 6 h., 2 Gy | 166 | 2 |
| GSE59861_6h_5cGy | - | 6 h., 5 cGy | 150 | 4 |
Here we present the results of the comparison of differentially expressed genes for doses 5 cGy and 2 Gy as a time series, grouped by time after exposure to IR (3, 6, 12 and 24 hours, Figure 1 and Figure 2, Table S1, Table S2).
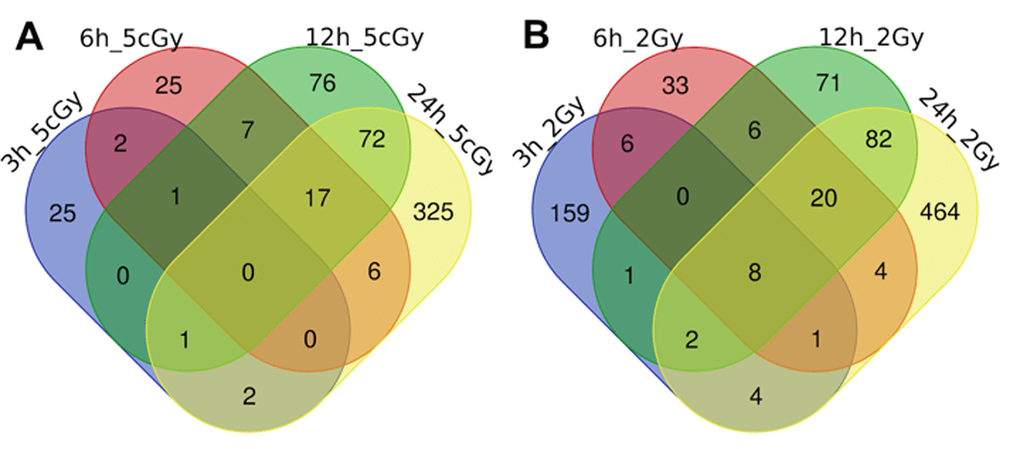
Figure 1. Venn diagrams, illustrating overlapping effects (up-regulated genes) of IR at different times of exposure: 3, 6, 12, 24 hours for the doses 5 cGy (A) and 2 Gy (B). Numbers indicate the amount of common/unique differentially expressed genes for the studied groups (Table S1).
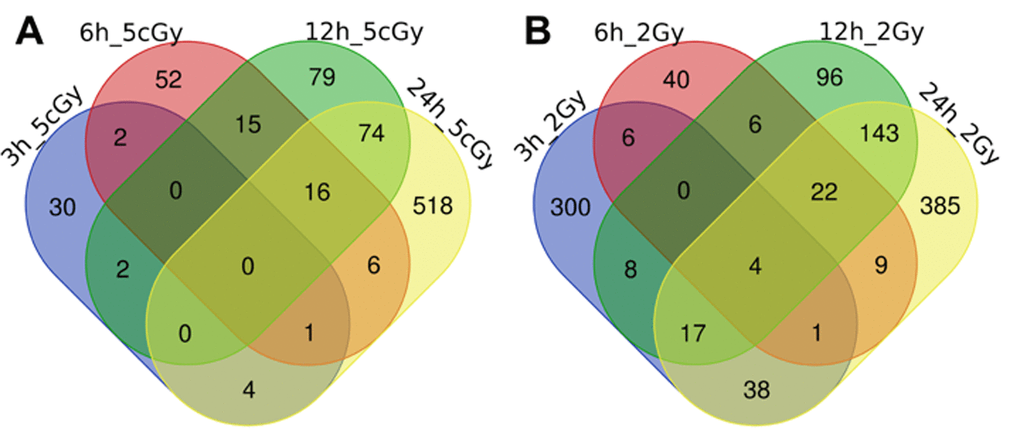
Figure 2. Venn diagrams, illustrating overlapping effects (down-regulated genes) of IR at different times of exposure: 3, 6, 12, 24 hours for the doses 5 cGy (A) and 2 Gy (B). Numbers indicate the amount of common/unique differentially expressed genes for the studied groups (Table S2).
Exposure of cells to 5 cGy of X-ray 3 hours after irradiation leads to the activation of the genes involved in connective tissue/extracellular matrix remodelling; in particular, we found upregulated genes responsible for lipids biosynthesis: ceramides, sphingolipids and phosphatidylinositol phosphate. Also, among the other up-regulated genes, we found genes responsible for the development of alveoli, as well as the FGF22, which plays a role in the regulation of cell growth and development [13]. Among down-regulated genes were CEP152, necessary for centriole assembly [14], and DYNLRB2, involved in the synthesis of new organelles and mitotic spindle organization [15].
According to GO analysis, 6 hours after X-ray exposure to 5 cGy, further processes occur in the cells: cell cycle arrest (IRF6, DDIT3) [16], apoptosis signalling and programmed cell death (DDIT3, DDIT4, TRAF1, SPATA2) [17,18], development of the cell response to ROS (SESN2, DDIT4) [19], arrest of the translation in the endoplasmic reticulum in response to cellular stress, DNA damage response (DDIT4), etc. At the same time there is a suppression of the expression of genes involved in cytokines productions (EGR1, BIRC3, HSPA1A, CCL1, LY9), MAP-kinase cascade (TAB2, DUSP1, DUSP2, KLHDC10), protein folding and their transport across the endoplasmic reticulum.
Signalling pathways activation analysis showed that proteasomal protein degradation and EGF-Rab5 (linked to endosomes internalisation and extracellular signalling) pathways are up-regulated [20] (Table S3). At the same time, protein folding and cytoskeleton development pathways are down-regulated (Table S4).
12 hours after exposure to 5 cGy of X-ray we still observe high expression level of genes involved in programmed cell death (DAB2IP, SFRP2, CHAC1, RRAGC, RFFL, DDIT4, OGT, C4ORF14, ATF4, KRT36, TRAF1), while DNA replication genes (CCNL1, CDC6, SLFN11, MSH6), ROS-responsive genes (DUSP1, FOS, PPIF), and genes allowing G1/S transition (CCNL1, CDC6, CCND1) are suppressed. For this time point, analyses demonstrate a strong up-regulation of the Wnt/Notch pathway, linked to apoptosis and the protein degradation processes [21] (Table S3).
Signalling pathways activation analysis showed that, even 24 hours after the exposure, the Wnt/Notch pathway is still hyper-activated. In addition, we observed an increased activation of a number of signaling pathways associated with syndecan (Table S3). Syndecans mediate cell adhesion, cytoskeleton reorganization, and cellular signaling due to their function as co-receptors for many ligands including FGF, VEGF, TGFβ and fibronectin [22]. In particular, syndecans play an important role in extracellular matrix remodelling and wound healing [23,24]. In accordance with that, we also observed up-regulation of pathways linked to the activation of EGFR, VEGFR1 and VEGFR2 receptors, the Hedgehog pathway (important for cell differentiation [25]), as well as pathways involved in the positive regulation of cell adhesion and glucose uptake (Table S3). Taken together, our data suggest that by 24 hours after exposure to 5 cGy of X-ray the processes of cellular degradation and programmed cell death are replaced by cellular regeneration.
Exposure of the fibroblasts to 2 Gy of X-ray 3 hours after irradiation results in the development of the classical irradiation response, up-regulation of POLH, CDKN1A, MTA1, GADD45A, AEN [26]: simultaneous expression of proteins responsible for cell cycle arrest CDKN1A, GADD45A and DNA repair, POLH. Pathway analysis shows that X-ray exposure induces p53-mediated DNA damage response, BRCA1-mediated cell cycle arrest and activates PI3K/Akt pathway. At the same time, pathways related to syndecans and Catenin Beta 1, and those involved in cell adhesion and proliferation are suppressed [27].
6 hours after exposure to 2 Gy of X-ray we still observe stress-response in the cells. In particular, apoptotic processes are induced: TRIAP1, PHLDA1, FAS, CDKN1A,TP53INP1, etc. Analysis shows up-regulation of genes responsible for cellular aging (senescence): TGFB3, DKK1, GCLC, KCNMB1, CDKN1A, DDC, MYST3 [28–30]. In parallel, cells activate defence mechanisms against ROS and DNA damage: glutathione is synthesized (CHAC1, GCLC и др.) [31], polymerase POLH and PCNA factor expression levels are enhanced.
12 hours after exposure we still observe signs of apoptosis: up-regulated expression of DAB2IP, SFRP2, ZMAT3, BCL6, TP53INP1, PHLDA3, DDIT4, TOP2A, BBC3, CDKN1A, APH1B, ATF4, SCIN. In parallel, an irradiation response is unfolding, similar to the one we saw 6 h after exposure: analysis shows activation of POLH, DDB2,TOP2A, responsible for double-stranded break (DSB) repair. At the same time, we observe a down-regulation of genes important for DNA replication (CCNE2, MCM10, GINS3, MCM3, etc.) [32], protein biosynthesis and G1/S transition (CCND1, CCNE2, etc.) [33]. Analysis of signalling pathways showed activation of the Wnt/Notch pathway, BRCA1 G1/S cell cycle arrest, p53-mediated DNA damage response, and the degradation of AKAP1 protein, which plays an important role in the regulation of protein kinase A function and its cellular distribution (Table S3) [34]. In particular, it has been shown that AKAP1 is necessary for the maintenance of mitochondrial homeostasis and, as a consequence, for the regulation of oxidative phosphorylation and senescence [35]. At the same time we observe down-regulation of the pre-replicative complex through the inhibition of the CDC6/ORC pathway (Table S4) [36] and the suppression of DNA single-strand breaks (SSB) repair processes.
24 hours after exposure to 2 Gy of X-ray we observe initiation of cell responses to ROS (SESN1, SESN2) [19] and cell starvation, leading to lipids breakdown (AKR1C3). We observe activation of catabolic processes, autophagy (TRIM22, TP53INP1) and suppression of transcription and translation. At the level of the signalling pathways, we observe strong (equal to the 12-hour level) deprivation of DNA replication, DNA SSB repair, and mitosis (G1/S transition, mitotic spindle formation, cyclins synthesis). On the other hand, pathways related to apoptosis (Notch/Wnt, ATM) and mitotic arrest are up-regulated (Table S3).
Comparison of gene expression of the fibroblasts exposed to different X-ray doses at the same time point after irradiation (Table S1 and Table S2) shows that: 3 hours after exposure there are just 11 commonly expressed genes (4 up- and 7 down-regulated), which complicates proper analysis of the common pathways; 6 hours after exposure there are 5 commonly up-regulated genes and no common down-regulated genes; 12 hours after irradiation for both doses we observe suppression of replication and transcription (KLF4, VEGFA, ZNF691, DAB2IP), and enhancement of p38-MAPK, Wnt and VEGF pathway activities (Table S3); 24 hours after exposure to X-ray, among common processes, we detect stimulation of hydrogen peroxide decomposition and glutathione synthesis (protection against ROS), while genes involved in DNA replication and G/S transition are suppressed, as well as gene expression in general.
Comparison of ageing with the effect of low and high doses of radiation on the cells
Comparison of the data on gene expression in fibroblasts 3 hours after exposure to 2 Gy X-ray with replicatively aged cells (data sets EMTAB2086_70_vs_30 and EMTAB2086_80_vs_30) shows that, among the common differentially expressed genes, there is an up-regulation of genes involved in DNA damage signalling through p53 pathway (BTG2, SESN2, CDKN1A, GADD45A), as well as the general stress response (EPHA2, BTG2, SESN2, SESN1, CDKN1A, EDEM3, GADD45A). Among commonly down-regulated ones, we found genes responsible for cell cycle progression (CNOT6, CDKN1B, PRKDC, MSH6); whereas gene TAOK2, involved in stress-response [37,38], is in fact up-regulated Table S2, Table S3, Figures 3, 4).
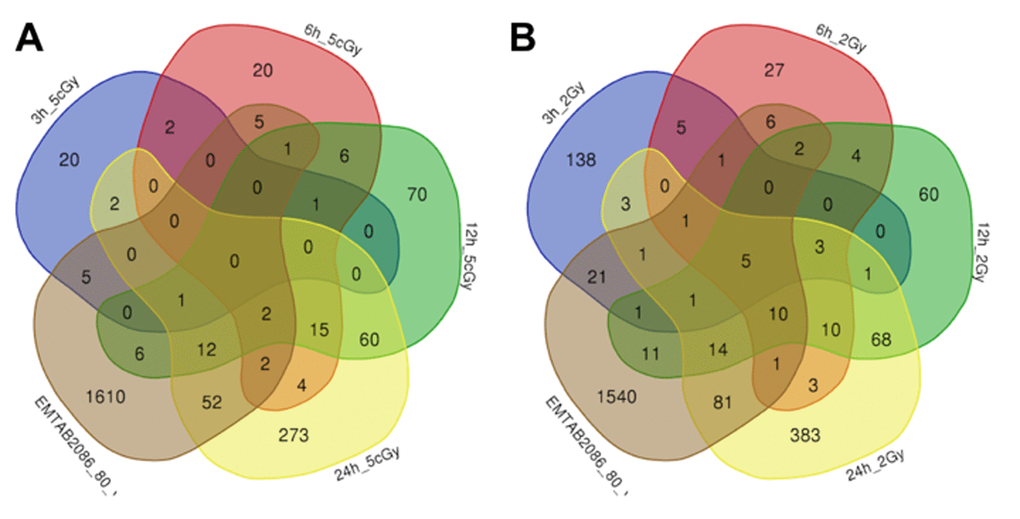
Figure 3. Venn diagrams, illustrating overlapping effects (up-regulated genes) of replicative aging and IR at different times of exposure: 3, 6, 12, 24 hours for the doses 5 cGy (A) and 2 Gy (B). Numbers indicate the amount of common/unique differentially expressed genes for the studied groups (Table S1).
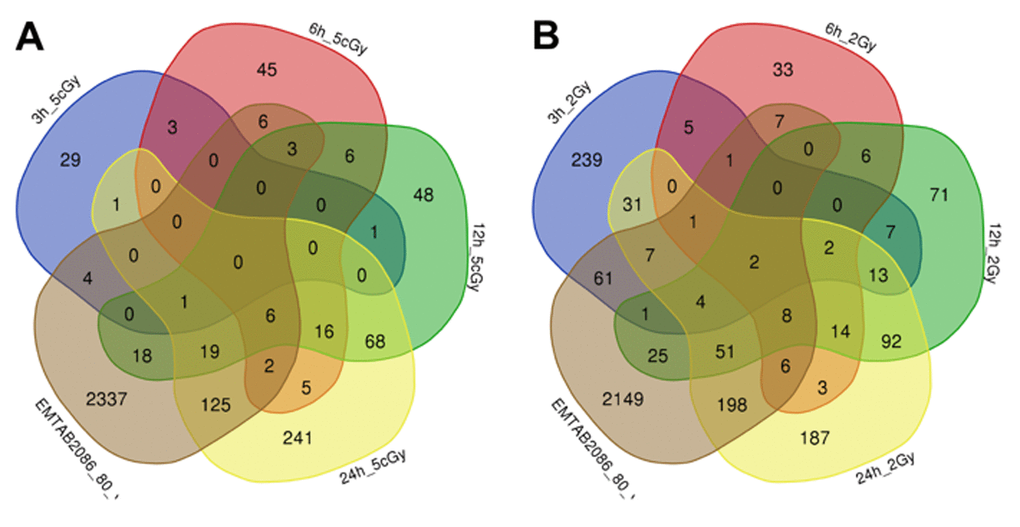
Figure 4. Venn diagrams, illustrating overlapping effects (down-regulated genes) of replicative aging and IR at different times of exposure: 3, 6, 12, 24 hours for the doses 5 cGy (A) and 2 Gy (B). Numbers indicate the amount of common/unique differentially expressed genes for the studied groups (Table S2).
Comparison of gene expression 6 hours after exposure to 2 Gy X-ray with replicatively aged cells show that both of them slow down cellular metabolism because of the lack of amino acids, so-called cellular starvation (SESN2, SESN1, FAS, CDKN1A), along with an initiation of apoptosis in response to external/internal signals (MDM2, BCL2L1, TP53INP1, CDKN1A, TNFRSF10B, FAS, BLOC1S2) and DNA damage signals. At this time point, down-regulated genes for 2 Gy are those involved in cleavage of intra-strand DNA crosslinks and DNA repair (RFWD3, ERCC3, HMGB1, FANCE, FANCL, MSH6); down-regulated genes for 5 cGy are those involved in cell proliferation (EGR1, TXNRD1, TCF7L2, MRPS27, HSPD1, EPS8).
Results of comparison of gene expression 12 hours after irradiation (2 Gy) with replicatively aged fibroblasts demonstrates suppression of cell p53-mediated DNA damage response; among common up-regulated we found genes involved in apoptosis and response to radiation (MDM2, BCL2L1, TP53INP1, CDKN1A). Common differentially expressed genes for replicatively aged cells and fibroblasts irradiated with 5 cGy are signals about external stimuli (EGR2, SESN2, GDF15). Down-regulated genes for 2 Gy are those involved in DNA replication (20 common genes), DNA repair and initiation of mitosis (7 and 13 common genes). Suppressed genes for 5 cGy are cell cycle genes.
Common differentially expressed genes for fibroblasts 24 hours after irradiation (5 cGy and 2 Gy doses) and fibroblasts from old patients, as well as replicatively aged cells are presented in Table S3 and Table S4. Comparison of gene expression 24 hours after irradiation (2 Gy) with data sets EMTAB2086_70_vs_30 and EMTAB2086_80_vs_30 reveals that there are some shared up-regulated genes: genes involved in cell death (17 genes), p53-mediated DNA damage response (9 genes), as well as genes involved in cell cycle arrest (5 genes) and autophagy (6 genes). Common downregulated genes (2 Gy) are those which play a role in mitosis (including replication and formation of mitotic spindle) and DNA repair. For smaller dosed (5 cGy) and replicatively aged cells, common responses include MAP-kinase signalling (4 genes), activation of other signalling cascades (5 genes), down-regulation of genes involved in DNA replication (over 30 genes), cell division (15 genes) and DNA DSB repair.
Signaling pathway analysis revealed the most relevant results for the differential expression/activation of signalling pathways in the cells 24 hours after exposure to IR (Table S3, Table S4). The results of the comparison of gene expression in replicatively old fibroblasts (data sets EMTAB2086_70_vs_30 and EMTAB2086_80_vs_30) and in cells exposed to 5 cGy of X-ray (24 hours after irradiation) demonstrate several common trends for both data sets, particularly an activation of signalling pathways important for cell cycle and adsorption of salts/ions, while comparison to the effects of 2 Gy 24 hours after exposure shows, among common activated pathways, G1/S cell cycle arrest and DNA damage response (BRCA1/E2 pathway).
We have not found any common differentially expressed genes in irradiated fibroblasts and fibroblasts obtained from middle-aged patients (GSE55118_middle_ vs_young), however there were some common changes when we compared them to “old” fibroblasts (GSE55118_old_vs_young). In particular, for the X-ray dose of 2 Gy we observe common down-regulation of pathways involved in cell division: chromosome segregation, centrosomes separation in mitosis, mitotic spindle formation, as well as GATA2-mediated transcription (Table S4). Among shared up-regulated pathways, when we compared “old” fibroblasts to those irradiated by the X-ray dose of 5 cGy, we observe the above-mentioned cell adhesion pathways, while the GATA2 pathway is down-regulated