Mutational analyses
NGS analysis revealed mutations in 19 different genes in 115 (92%) patients. Among these 115 patients, 34 (27.2%) had mutations in a single gene, 35 (28%) had mutations in two genes, 28 (22.4%) had mutations in three genes, 17 (13.6%) had mutations in four genes, and one had mutations in six genes. The most frequently mutated gene was TET2 (26.4%). The remaining mutated genes in the order of decreasing frequency were: ASXL1, 24.8%; FLT3, 23.2%; NPM1, 21.6%; DNMT3A, 19.2%; NRAS, 17.6%; CEBPA, 14.4%; IDH2, 12%; TP53, 12%; RUNX1, 8.8%; IDH1, 8.0%; and KIT, 5.6%; mutations in ETV6, SRSF2, U2AF1, PHF6, EZH2, CBL and CSF3R were rare (<5.0%); mutations were not identified in KRAS, JAK2, ZRSR2 and GATA2 genes (Figure 1A, Table 2). In comparison to patients with complex karyotypes, mutations in DNMT3A (21.7% vs. 0, P= 0.0341), TET2 (34.8% vs. 10.5%, P= 0.0485), NPM1 (14.5% vs. 0, P= 0.0047) and FLT3 (23.2% vs. 0, P= 0.0184) were more common in patients with normal karyotype. Conversely, the rate of TP53 mutations was significantly higher in patients with complex karyotypes than those with the normal karyotype (57.9% vs. 2.9%, P< 0.0001; Table 2). Ten out of fifteen patients (66.7%) with TP53 mutations belonged to the t-AML or the AML-MRC group. We divided the patients based on their age into three subgroups (60-66, 67-74 and 75-86 years) using ROC curve analysis. Mutations in IDH1 were more common in patients aged 67–74 years than those aged 60–66 years (18.0% vs. 3.1%, P=0.0248; Figure 1B). The rate of RUNX1 mutations were significantly higher in patients aged 75–86 years than those aged 60-66 year age group (14.3% vs. 4.6%, P= 0.0264; Figure 1B).
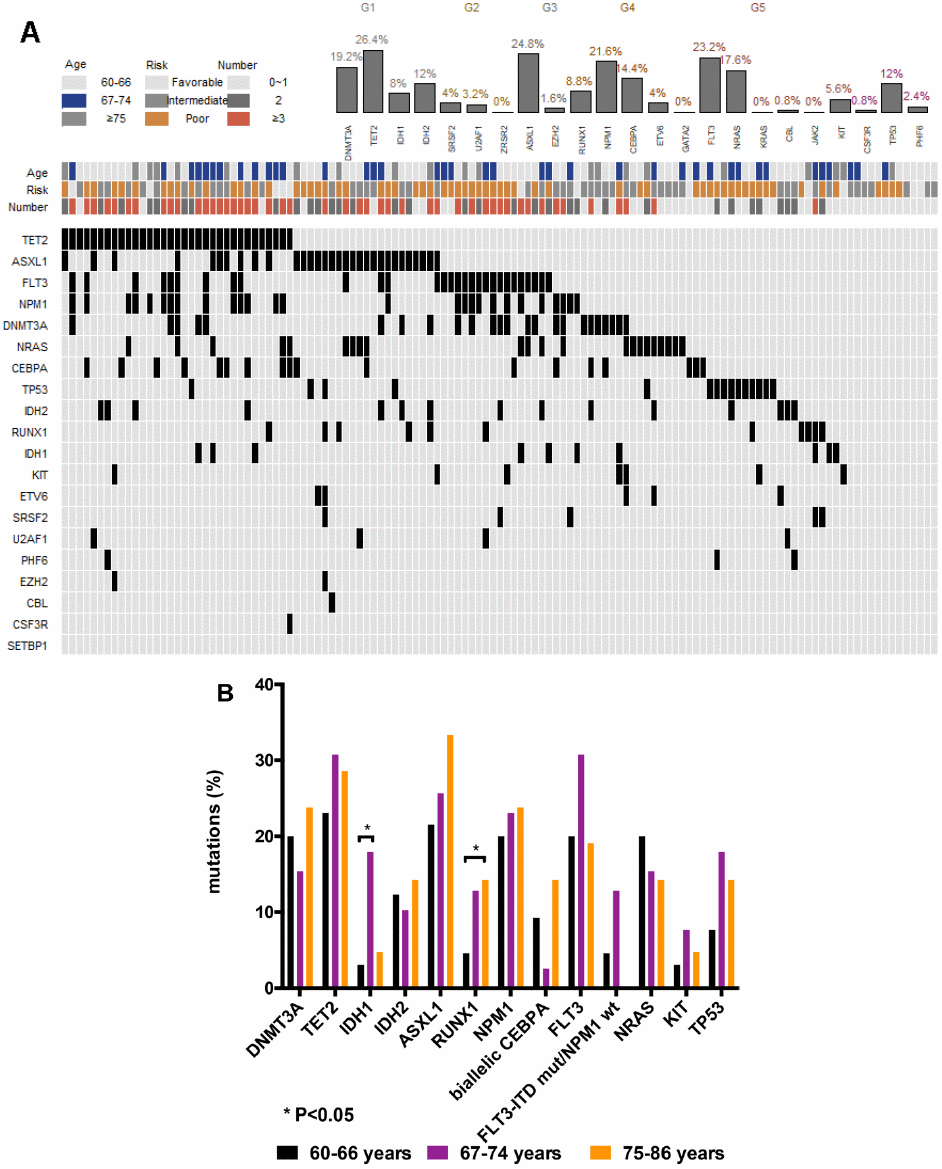
Figure 1. Genetic landscape of elderly AML patients. (A) Heatmap showing associations between different gene mutations. Each column represents one patient. (B) Gene mutations in 125 AML patients ≥ 60 years of age at primary diagnosis. Bar chart showing the 13 most commonly mutated genes in elderly AML patients aged 60-66, 67-74, and 75-86 years at primary diagnosis.
Table 2. Mutations.
| Mutations | n (%) | Normal cytogenetic (n=69) n (%) | Complex karyotypes (n=19) n (%) | P |
| DNA methylation | DNMT3A | 24 (19.2) | 15 (21.7) | 0 | 0.0341 |
| TET2 | 33 (26.4) | 24 (34.8) | 2 (10.5) | 0.0485 |
| IDH1 | 10 (8) | 6 (8.7) | 0 | 0.3332 |
| IDH2 | 15 (12) | 12 (17.4) | 0 | 0.0619 |
| RNA splicing | SRSF2 | 5 (4) | 0 | 0 | |
| U2AF1 | 4 (3.2) | 0 | 0 | |
| ZRSR2 | 0 | 0 | 0 | |
| Epigenetic modifiers | ASXL1 | 31 (24.8) | 16 (23.2) | 6 (31.6) | 0.5509 |
| EZH2 | 2 (1.6) | 0 | 0 | |
| Transcription factors | RUNX1 | 11 (8.8) | 6 (8.7) | 0 | 0.3332 |
| NPM1 | 27 (21.6) | 21 (14.5) | 0 | 0.0047 |
| CEBPA | 18 (14.4) | 11 (15.9) | 2 (10.5) | 0.7256 |
| biallelic CEBPA | 10 (8) | 6 (8.7) | 4 (22.2) | 0.0882 |
| ETV6 | 5 (4) | 3 (4.3) | 0 | |
| GATA2 | 0 | 0 | 0 | |
| Activited signaling | FLT3 | 29 (23.2) | 16 (23.2) | 0 | 0.0184 |
| FLT3-ITD | 20 (16) | 13 (18.8) | 0 | 0.0624 |
| FLT3-TKD | 11 (8.8) | 3 (4.3) | 0 | |
| NRAS | 22 (17.6) | 16 (23.2) | 1 (5.3) | 0.1054 |
| KRAS | 0 | 0 | 0 | |
| CBL | 1 (0.8) | 0 | 1 (5.3) | |
| JAK2 | 0 | 0 | 0 | |
| KIT | 7 (5.6) | 1 (1.4) | 1 (5.3) | |
| Tumor suppressors | CSF3R | 1 (0.8) | 0 | 0 | |
| TP53 | 15 (12) | 2 (2.9) | 11 (57.9) | <0.0001 |
| PHF6 | 3 (2.4) | 0 | 0 | |
Association between cytogenetics, gene mutations and clinical outcomes
After a median follow-up of 12 months (range: 2-82 months), 86 (68.8%) and 94 patients (75.2%) treated with D-CAG achieved CR after one cycle and two cycles of induction, respectively. Patients in the low- or intermediate-risk groups showed a higher CR rate than patients in the high-risk group, but the differences were not statistically significant (91.7% vs. 78.4% vs. 69.4%, P= 0.2053; Table 3).
Table 3. Response and clinical outcome.
| Clinical features | CR n (%) | Median OS (months) | Median DFS (months) | 1-year OS (%) | 2-year OS (%) |
| All patients (n=125) | 94 (75.2) | 16 | 12 | 59.8 | 36.5 |
| Age (years) | P=0.5473 | P=0.0070 | P=0.1220 | | |
| 60-66 (n=65) | 51 (78.5) | 19 | 15 | 70.6 | 45.1 |
| 67-74 (n=39) | 29 (74.4) | 14 | 11 | 53.2 | 36.6 |
| 75-86 (n=21) | 14 (66.7) | 9 | 7 | 40.6 | 20.3 |
| Patients aged 60-66 years (n=65) | P=0.3612 | P=0.0134 | P=0.4344 | | |
| Low- and intermediate-risk (n=37) | 31 (83.8) | 28 | 15 | 84.0 | 53.7 |
| High-risk (n=28) | 20 (71.4) | 13 | 14 | 52.8 | 35.2 |
| Patients aged 67-74 years (n=39) | P=1.0000 | P=0.0681 | P=0.0014 | | |
| Low- and intermediate-risk (n=17) | 13 (76.5) | 32 | 19 | 69.0 | 40.0 |
| High-risk (n=22) | 16 (72.7) | 12 | 6 | 52.6 | 22.8 |
| Patients aged 75-86 years (n=21) | P=0.6557 | P=0.2110 | P=0.1048 | | |
| Low- and intermediate-risk (n=8) | 6 (75.0) | 18 | 13 | 19.4 | N/A |
| High-risk (n=13) | 8 (61.5) | 9 | 6 | 10.3 | N/A |
| Numbers of mutations | P=0.0035 | P=0.1570 | P=0.6556 | | |
| 0-1 (n=44) | 26 (59.1) | 12 | 18 | 47.0 | 26.9 |
| 2 (n=35) | 32 (91.4) | 18 | 11 | 69.6 | 39.3 |
| ≥3 (n=46) | 36 (78.3) | 17 | 14 | 64.2 | 34.7 |
| DNMT3A | P=0.1208 | P=0.6243 | P=0.7438 | | |
| mut (n=24) | 15 (62.5) | 13 | 13 | 53.0 | 26.5 |
| wt (n=101) | 79 (78.2) | 16 | 12 | 61.4 | 39.1 |
| TET2 | P=1632 | P=0.6365 | P=0.6875 | | |
| mut (n=33) | 28 (84.8) | 17 | 13 | 65.6 | 38.6 |
| wt (n=92) | 66 (71.7) | 15 | 11 | 57.7 | 37.3 |
| IDH1 | P=0.7028 | P=0.8188 | P=0.2503 | | |
| mut (n=10) | 7 (70) | Undefined | Undefined | 51.4 | N/A |
| wt (n=115) | 87 (75.7) | 16 | 12 | 60.5 | 37.6 |
| IDH2 | P=0.7603 | P=0.1423 | P=0.8574 | | |
| mut (n=15) | 12 (80) | 20 | 13 | 86.7 | 40.5 |
| wt (n=110) | 82 (74.5) | 15 | 11 | 55.7 | 37.3 |
| ASXL1 | P=1.0000 | P=0.4425 | P=0.0801 | | |
| mut (n=31) | 24 (77.4) | 16 | 8 | 56.5 | 31.4 |
| wt (n=94) | 70 (74.5) | 16 | 14 | 60.6 | 39.7 |
| RUNX1 | P=1.0000 | P=0.7141 | P=0.4392 | | |
| mut (n=11) | 8 (72.7) | 18 | 18 | 71.6 | 42.9 |
| wt (n=114) | 86 (75.4) | 16 | 12 | 58.7 | 37.5 |
| NPM1 | P=0.4602 | P=0.3479 | P=0.1092 | | |
| mut (n=27) | 22 (81.5) | 19 | 18 | 65.6 | 41.4 |
| wt (n=98) | 72 (73.5) | 15 | 10 | 57.8 | 36.8 |
| CEBPA | P=0.3451 | P=0.2662 | P=0.0994 | | |
| biallelic mut (n=10) | 9 (90) | 15.5 | 9.5 | 60.0 | 20.0 |
| monoallelic mut (n=8) | 7 (87.5) | 19 | 18 | 75.0 | 46.9 |
| wt (n=107) | 78 (72.9) | 16 | 12 | 58.8 | 39.9 |
| FLT3 | P=0.8065 | P=0.2247 | P=0.8653 | | |
| mut (n=29) | 21 (72.4) | 12 | 18 | 46.8 | 32.8 |
| wt (n=96) | 73 (76.0) | 17 | 12 | 63.3 | 37.2 |
| NRAS | P=1.0000 | P=0.7962 | P=0.7377 | | |
| mut (n=22) | 17 (77.3) | 28 | 19 | 58.7 | 52.8 |
| wt (n=103) | 77 (74.8) | 15 | 11 | 60.0 | 34.9 |
| KIT | P=1.0000 | P=0.4152 | P=0.1337 | | |
| mut (n=7) | 5 (71.4) | 14 | 7.5 | 60.0 | N/A |
| wt (n=118) | 89 (75.4) | 16 | 12 | 59.7 | 39.2 |
| TP53 | P=1.0000 | P=0.0657 | P=0.0649 | | |
| mut (n=15) | 11 (73.3) | 10 | 7 | 46.7 | 15.6 |
| wt (n=110) | 83 (75.5) | 18 | 14 | 66.3 | 39.7 |
| Cytogenetics | | | | | |
| P=0.5641 | P=0.0041 | P=0.0001 | | |
| complex karotypes (n=19) | 13 (68.4) | 9 | 5 | 35.5 | 11.8 |
| others (n=106) | 81 (76.4) | 19 | 15 | 64.6 | 41.5 |
| P=0.7713 | P<0.0001 | P<0.0001 | | |
| -5/5q-, -7/7q- (n=18) | 13 (72.2) | 8.5 | 6 | 27.8 | 5.6 |
| others (n=107) | 81 (75.7) | 19 | 15 | 65.8 | 44.5 |
| P=0.2911 | P=0.3143 | P=0.0004 | | |
| monosomal (n=12) | 11 (91.7) | 13 | 5 | 57.1 | 19.0 |
| others (n=113) | 83 (73.5) | 17 | 14 | 60.1 | 40.3 |
| risk status | P=0.3727 | P=0.0022 | P=0.0041 | | |
| Low-risk (n=12) | 11 (91.7) | Undefined | 15 | 88.9 | 71.1 |
| Intermediate-risk (n=50) | 39 (78.0) | 20 | 15 | 74.0 | 47.9 |
| High-risk (n=63) | 44 (69.8) | 11 | 8 | 44.1 | 25.8 |
| Abbreviations: CR: complete remission; OS: overall survival; DFS: disease-free survival; mut: mutated status; wt: wild type. |
At the final analysis on July 30, 2019, the median OS and DFS of all patients were 16 and 12 months, respectively. The median OS was 19, 14 and 9 months for patients aged 60–66 years, 67–74 years, and ≥75 years, respectively (P=0.007, Figure 3). The one-year and 2-year OS rates were 59.8% and 36.5%, respectively. The one-year and 2-year DFS rates were 49.3% and 28.2%, respectively. Patients aged 60–66 years showed significantly longer OS than patients ≥75 years (median OS: 19 months vs. 9 month, P= 0.0042, Table 3, Figure 3). However, DFS was statistically similar for both groups (median DFS: 15 months vs. 7 months, P= 0.0526, Table 3). The OS and DFS rates were significantly longer in patients belonging to the low- and intermediate-risk groups than in patients belonging to the high-risk group (median OS: Undefined vs. 20 months vs. 11 months, P= 0.0022; median DFS: 15 months vs. 15 months vs. 8 months, P= 0.0041, respectively; Table 3, Figure 4). Patients with complex karyotypes showed significantly shorter median OS and DFS compared to patients with non-complex karyotypes (n=19; median OS: 9 months vs. 19 months, P= 0.0041; median DFS: 5 months vs. 15 months, P= 0.0001, respectively; Table 3, Figure 5). Moreover, patients with abnormalities in chromosomes 5 and/or 7 (-5/5q- and/or -7/7q-) showed significantly shorter median OS and DFS compared to patients without these abnormalities (-5/5q- and/or -7/7q-, n=18; median OS: 8.5 months vs. 19 months, P< 0.0001; median DFS: 6 months vs. 15 months, P< 0.0001, respectively; Table 3, Figure 5). Patients with monosomal karyotypes (n=12) showed similar median OS, but significantly shorter median DFS compared to other patients (median OS: 13 months vs. 17 months, P= 0.3143; median DFS: 5 months vs. 14 months, P= 0.0004; Table 3, Figure 5).
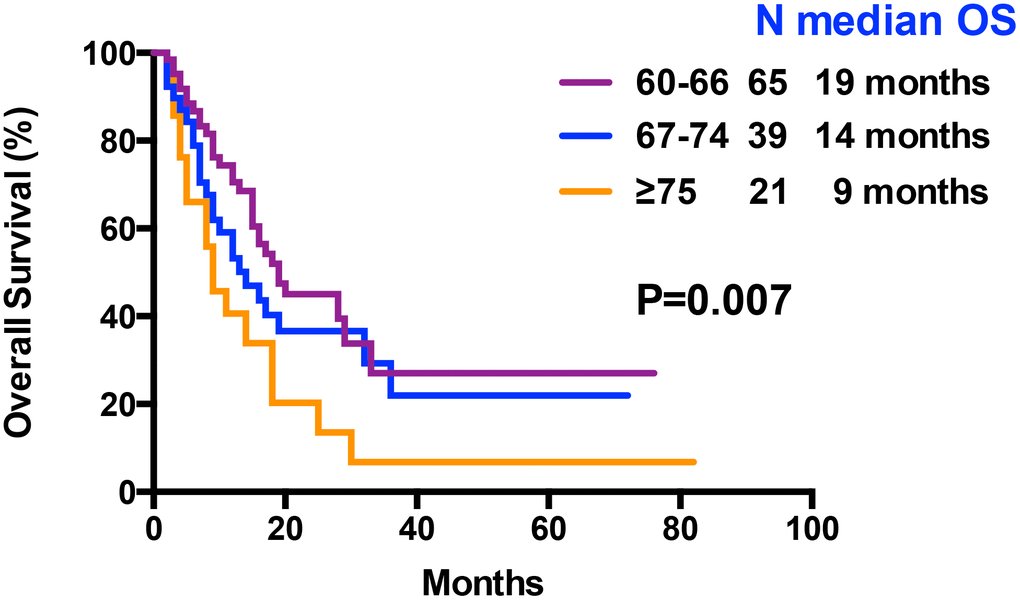
Figure 3. Kaplan–Meier curves associated with overall survival within age arms (60-66 vs 67-74 vs ≥ 75 years).
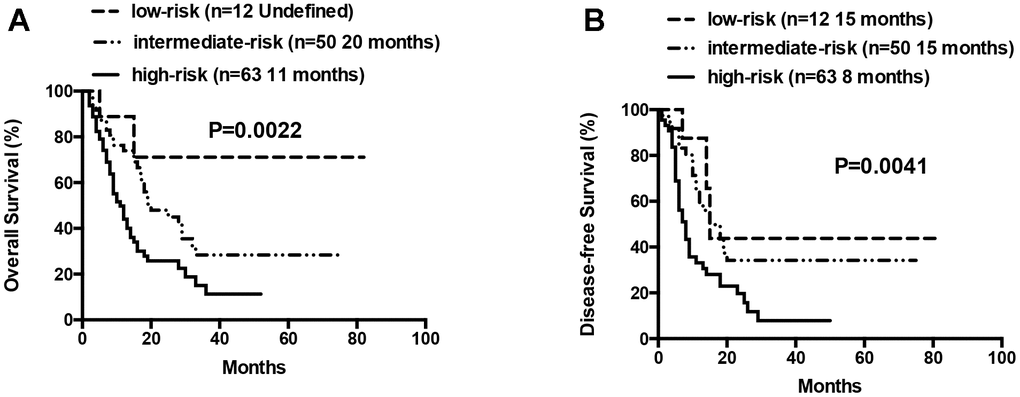
Figure 4. Overall survival and disease free survival according to risk groups (low-risk vs intermediate-risk vs high-risk). (A) Overall survival in low-, intermediate- and high-risk patients. (B) Disease free survival in low-, intermediate- and high-risk patients.
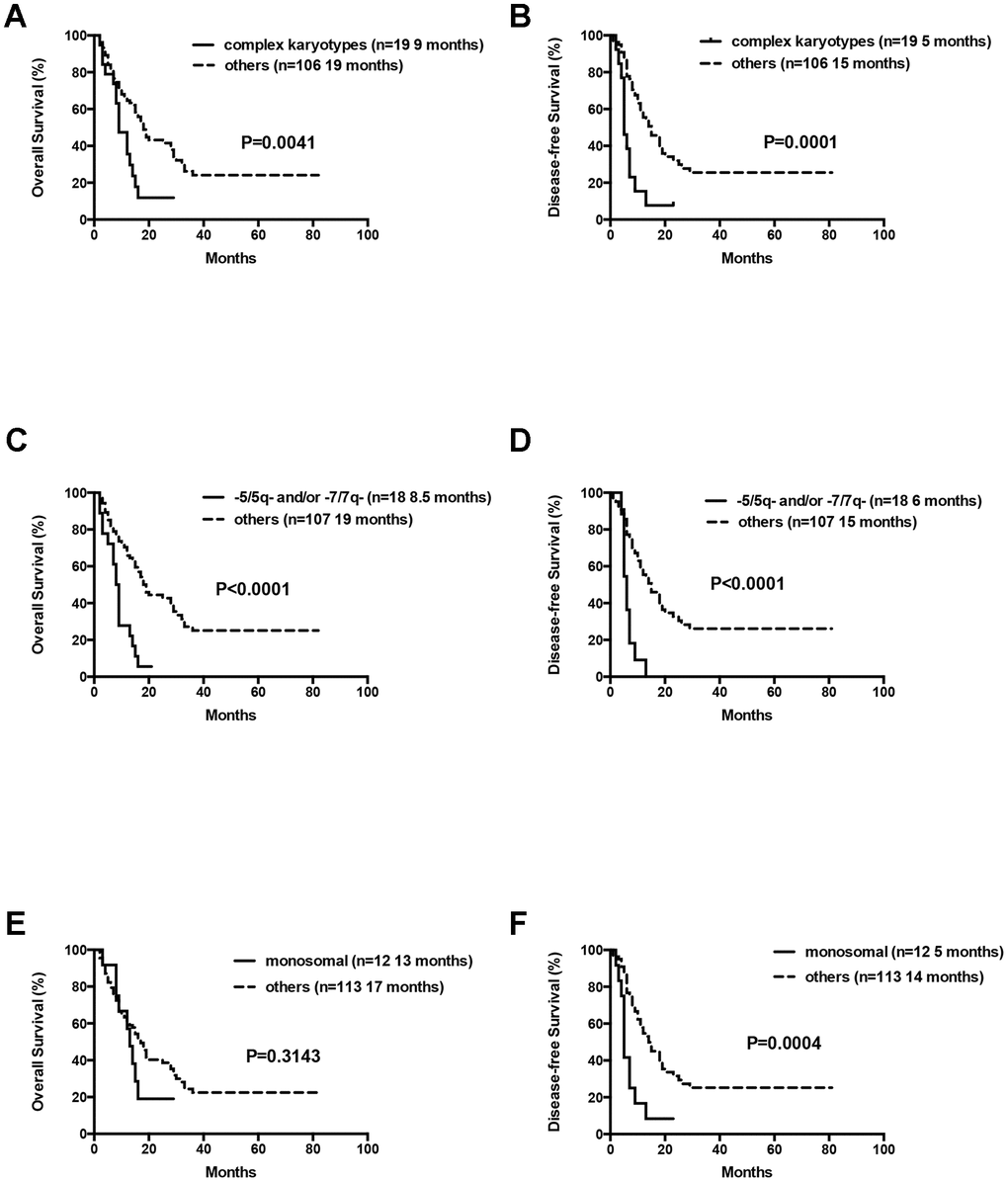
Figure 5. Overall survival and disease free survival according to cytogenetics. (A) Overall survival in AML patients with complex cytogenetics compared to others. (B) Disease free survival in AML patients with complex cytogenetics compared to others. (C) Overall survival in AML patients with abnormalities in -5/5q- and/or -7/7q- chromosomal deletions compared to others. (D) Disease free survival in AML patients with abnormalities in -5/5q- and/or -7/7q- chromosomal deletions compared to others. (E) Overall survival in AML patients with monosomal karyotype compared to others. (F) Disease free survival in AML patients with monosomal karyotype compared to others.
Patients in the 60-66 and 67-74 age groups showed better OS and DFS for the low- and intermediate-risk group patients compared to the high-risk group patients. But, these differences were not statistically significant except that median DFS for the low- and intermediate-risk groups was significantly longer than the high-risk group for patients aged 67-74 years (median DFS: 19 months vs. 6 months, P= 0.0014; Figure 6).
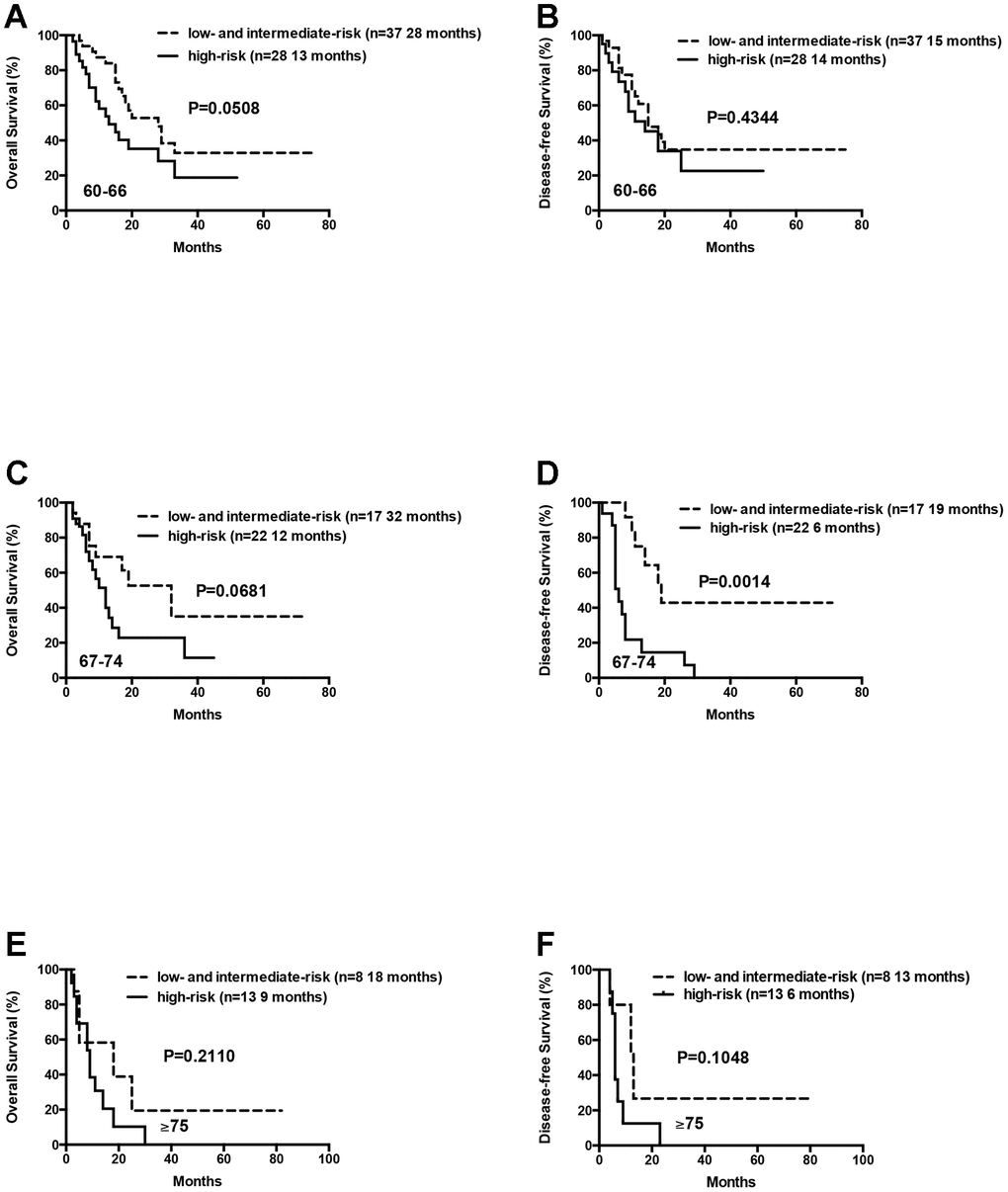
Figure 6. Overall survival and disease free survival of the AML patients according to risk groups (favorable and intermediate vs poor) within age arms (60-66 vs 67-74 vs ≥ 75 years). (A) Overall survival of the AML patients aged 60-66 years according to risk groups. (B) Disease free survival of the AML patients aged 60-66 years according to risk groups. (C) Overall survival of the AML patients aged 67-74 years according to risk groups. (D) Disease free survival of the AML patients aged 67-74 years according to risk groups. (E) Overall survival of the AML patients aged ≥75 years according to risk groups. (F) Disease free survival of the AML patients aged ≥75 years according to risk groups.
We also analyzed 21 patients who were ≥75 years, including 8 low- or intermediate-risk patients and 13 high-risk patients. The median OS and DFS were relatively longer in the low- and intermediate-risk group compared to the high-risk group, but the differences were not statistically significant (median OS: 18 months vs. 9 months, P= 0.2110; median DFS: 13 months vs. 6 months, P= 0.1048; Figure 6). This suggests that D-CAG is feasible for the treatment of AML patients above 75 years, especially those harboring high-risk karyotypes and genetic mutations.
The median OS for patients with 0 or 1, 2 or ≥3 gene mutations was not statistically significant (12 months vs. 18 months vs. 17 months, P= 0.1570). Moreover, OS and DFS for patients with or without mutations in genes such as ASXL1 and RUNX1 were similar (Table 3). Patients with wild-type TP53 (n=110) showed relatively longer OS and DFS compared to patients harboring TP53 mutations (n=15), but the differences were not statistically significant (median OS: 18 months vs. 10 months, P= 0.0657; median DFS: 14 months vs. 7 months, P= 0.0649; Table 3, Figure 7). Patients with TP53 mutations (VAF <20%) showed relatively longer survival than patients with TP53 mutations (VAF ≥20%), but, the data was not statistically significant after excluding two patients with low VAF (<20%) TP53 mutations (median OS: 18 months vs. 9 months, P= 0.1047; median DFS: 14 months vs. 7 months, P= 0.0511). Ten out of 15 patients with TP53 mutations were associated with -5/5q- and/or -7/7q- chromosomal deletions, whereas, the remaining 5 patients had isolated TP53 mutations. The patients with isolated TP53 mutations showed relatively longer median OS and DFS compared to patients with TP53 mutations and concomitant -5/5q- and/or -7/7q- chromosomal deletions (median OS: undefined vs. 9 months, P= 0.0740; median DFS: 15 months vs. 7 months, P= 0.3662; Table 3, Figure 7). The median OS and DFS was comparatively similar for patients with wild-type and isolated TP53 mutations (median OS: 18 months vs. undefined, P= 0.5146; median DFS: 14 months vs. 15 months, P= 0.7177; Table 3, Figure 7), but, significantly lower in patients with TP53 mutations and concomitant -5/5q- and/or -7/7q- chromosomal deletions (median OS: 9 months vs. 18 months, P= 0.0016; median DFS: 7 months vs. 14 months, P= 0.0023, respectively; Table 3, Figure 7). The median OS and DFS was statistically similar for AML patients with or without FLT3 mutations (median OS: 12 months vs. 17 months, P= 0.2247; median DFS: 18 months vs. 12 months, P= 0.8653, respectively; Table 3, Figure 8). Patients with FLT3-ITD mutations in the absence of NPM1 mutations (n=8) and the other patients (n=117) showed statistically similar median OS and DFS (median OS: 12 months vs. 16 months, P= 0.3967; median DFS: 25 months vs. 12 months, P= 0.8556, Figure 8).
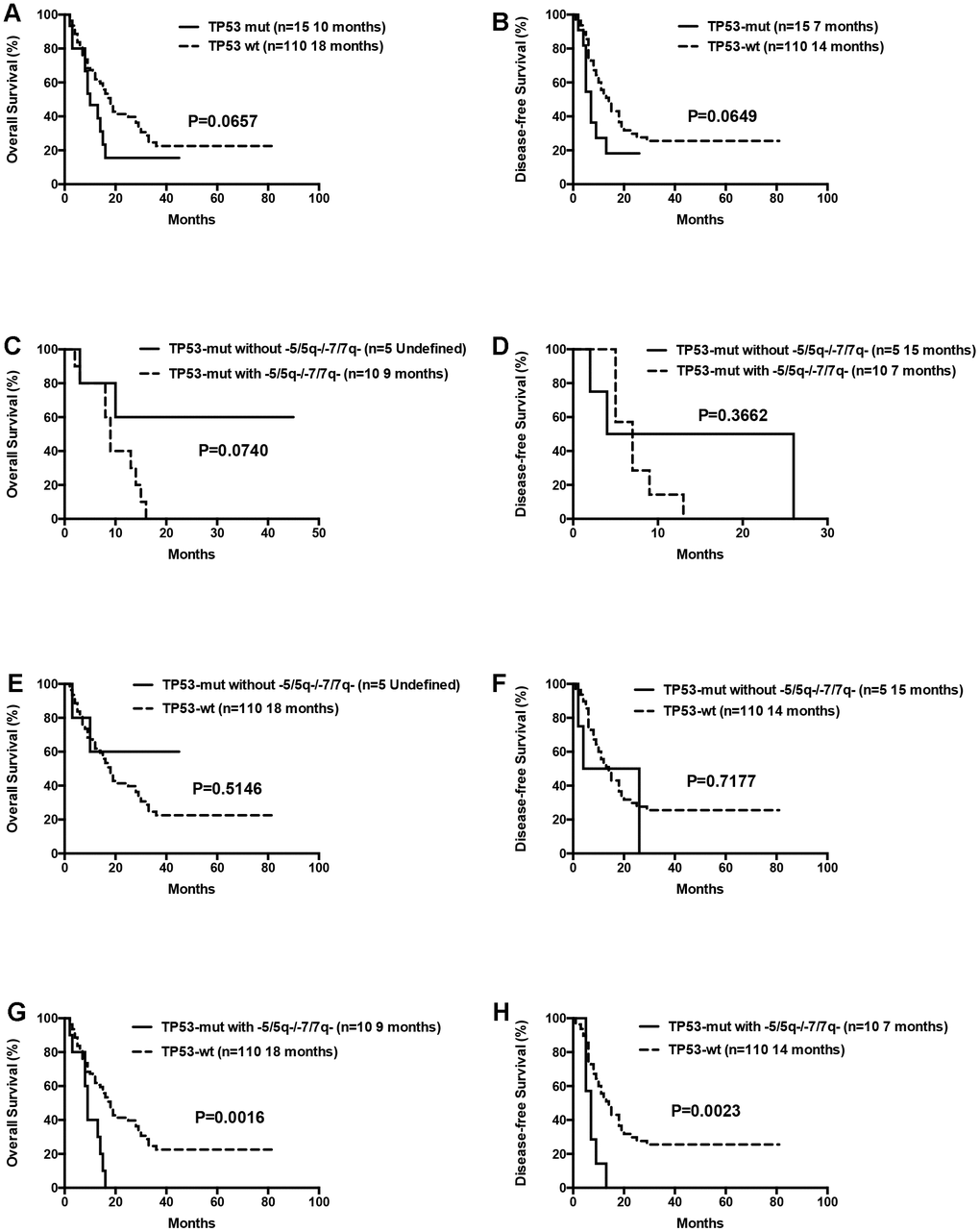
Figure 7. Overall survival and disease free survival according to TP53 mutations. (A) Overall survival in TP53 mutated compared to TP53 wild-type patients. (B) Disease free survival in TP53 mutated compared to TP53 wild-type patients. (C) Overall survival in isolated TP53 mutated patients compared to those with TP53 mutations and concomitant -5/5q- and/or -7/7q- chromosomal deletions. (D) Disease free survival in isolated TP53 mutated patients compared to those with TP53 mutations and concomitant -5/5q- and/or -7/7q- chromosomal deletions. (E) Overall survival in isolated TP53 mutated compared to TP53 wild-type patients. (F) Disease free survival in isolated TP53 mutated compared to TP53 wild-type patients. (G) Overall survival in patients with TP53 mutations and concomitant -5/5q- and/or -7/7q- chromosomal deletions compared to TP53 wild-type patients. (H) Disease free survival in patients with TP53 mutations and concomitant -5/5q- and/or -7/7q- chromosomal deletions compared to TP53 wild-type patients.
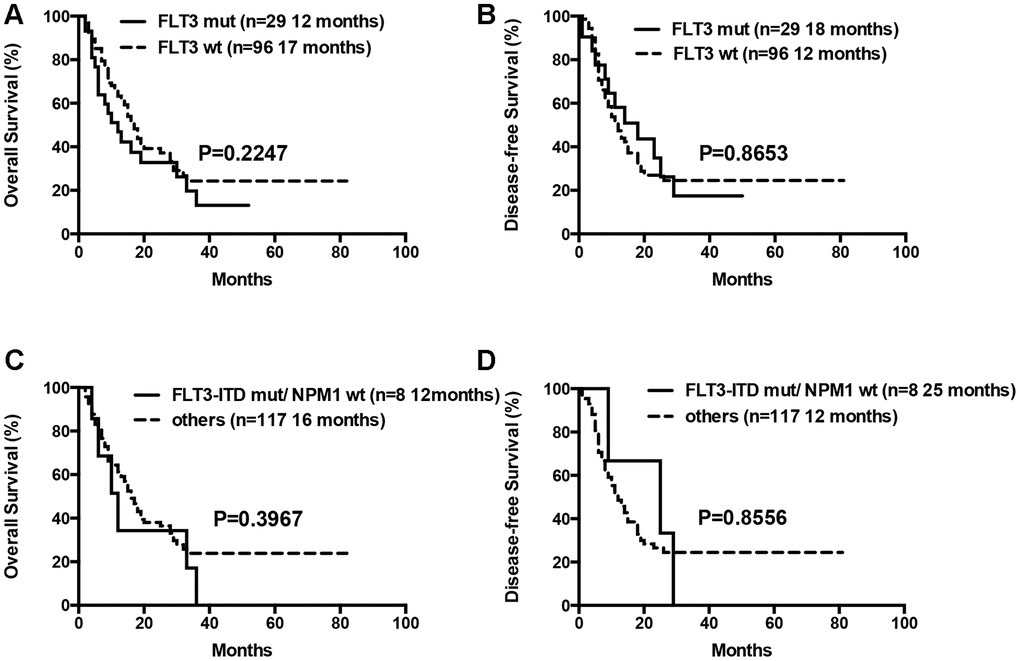
Figure 8. Overall survival and disease free survival according to FLT3 mutations. (A) Overall survival in FLT3 mutated compared to FLT3 wild-type patients. (B) Disease free survival in FLT3 mutated compared to FLT3 wild-type patients. (C) Overall survival in patients with FLT3-ITD mutations in the absence of NPM1 mutations compared to others. (D) Disease free survival in patients with FLT3-ITD mutations in the absence of NPM1 mutations compared to others.
Univariable analyses showed that age (≥75 years), complex karyotypes, -5/5q- and/or -7/7q- chromosomal deletions, and high-risk status were independent prognostic factors associated with shorter OS (Table 4). Factors such as monosomal karyotypes, total number of gene mutations, mutated TP53 and FLT3, and FLT3-ITD mutations in the absence of NPM1 mutations did not show prognostic significance in the univariable analysis. Multivariate analysis showed that age (over 75 years), high-risk status, and -5/5q- and/or -7/7q- chromosomal deletions were significant variables that predicted poor prognosis or decreased OS (Table 4).
Table 4. Univariate and multivariate analysis.
| Variable | Univariate analysis | Multivariate analysis |
| HR (95% CI) | P | HR (95% CI) | P |
| OS | | | | |
| age (≥75 yrs vs <75 yrs) | 1.995 (1.159~3.435) | 0.013 | 1.901 (1.099~3.288) | 0.022 |
| complex karotypes | 2.209 (1.254~3.893) | 0.006 | 0.689 (0.252~1.887) | 0.469 |
| -5/5q- and/or -7/7q- | 3.268 (1.855~5.757) | <0.001 | 3.206 (1.157~8.885) | 0.025 |
| monosomal karyotypes | 1.421 (0.704~2.871) | 0.327 | N/A | N/A |
| risk status | 1.967 (1.310~2.952) | 0.001 | 1.620 (1.058~2.482) | 0.027 |
| numbers of mutations (0-1) | N/A | 0.174 | N/A | N/A |
| numbers of mutations (2) | 1.461 (0.863~2.473) | 0.158 | N/A | N/A |
| numbers of mutations (≥3) | 0.889 (0.496~1.593) | 0.693 | N/A | N/A |
| Mutations | | | | |
| DNMT3A | 1.148 (0.652~2.021) | 0.631 | N/A | N/A |
| TET2 | 1.125 (0.717~1.764) | 0.609 | N/A | N/A |
| IDH1 | 0.891 (0.325~2.442) | 0.822 | N/A | N/A |
| IDH2 | 0.587 (0.282~1.224) | 0.155 | N/A | N/A |
| ASXL1 | 1.217 (0.729~2.032) | 0.453 | N/A | N/A |
| RUNX1 | 0.858 (0.373~1.977) | 0.72 | N/A | N/A |
| NPM1 | 0.767 (0.435~1.352) | 0.359 | N/A | N/A |
| CEBPA | 0.959 (0.527~1.745) | 0.892 | N/A | N/A |
| FLT3 | 1.370 (0.814~2.306) | 0.236 | N/A | N/A |
| FLT3-ITD mut/NPM1 wt | 1.424 (0.616~3.292) | 0.408 | N/A | N/A |
| NRAS | 0.926 (0.509~1.684) | 0.8 | N/A | N/A |
| KIT | 1.507 (0.547~4.151) | 0.428 | N/A | N/A |
| TP53 | 1.759 (0.944~3.277) | 0.075 | N/A | N/A |
| Abbreviations: OS: overall survival; HR: hazard ratio. |