Downregulation of KIF2C and TEKT2 is associated with male infertility and testicular carcinoma
Abstract
Background: Genetic factors are important in spermatogenesis and fertility maintenance, and are potentially significant biomarkers for the early detection of infertility. However, further understanding of these biological processes is required.
Methods: In the present study, we sought to identify associated genes by reanalyzing separate studies from Gene Expression Omnibus datasets (GSE45885, GSE45887 and GSE9210) and validation datasets (GSE4797, 145467, 108886, 6872). The differential genes were used the limma package in R language. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed by the clusterprofier package. The protein-protein interaction network was constructed by the STRING database. The interaction between mRNA and TF was predicted by miRWalk web. At last, The Cancer Genome Atlas data were used to identify hub gene expression levels in GEPIA web.
Results: The results showed that 27 shared genes associated with spermatogenesis. We effectively screen out two genes (KIF2C and TEKT2) and both validated by GSE4797, 145467, 108886 and 6872. Among 27 shared genes, KIF2C and TEKT2 both down-regulated in spermatogenesis. The network of TF-miRNA-target gene was established, we found KIF2C-miRNAs (has-miR-3154, 6075, 6760-5p, 1251-5p, 186-sp)-TFs (EP300, SP1) might work in spermatogenesis.
Conclusions: Our study might help to improve our understanding of the mechanisms in spermatogenesis and provide diagnostic biomarkers and therapeutics targets.
Introduction
Infertility, a couple's inability to have children after one year of normal sexual intercourse without protection, affects 10-15% of couples [1–4]. From the latest WHO statistics, nearly 50–80 million persons suffer from infertility [5, 6]. A few studies demonstrate that nearly 50% of all cases of infertility occur because of female factors, 20%–30% male factors, and 20%–30% couples [6–8]. Male infertility is a multifaceted and multiphenotypic disease, which affects about 7% of men worldwide [9]. Male infertility is a complex and heterogeneous phenotypic disease, from complete absence of sperm in testis to changes in sperm quality [10, 11]. Genetic factors account for at least 15% of male infertility. The three main causes of male infertility: spermatogenic function defects; ductal obstruction or disorder; hypothalamic–pituitary axis disorder. A patient with azoospermia is at the highest risk of carrying genetic abnormalities.
With the biotechnology’s improving, we can immediately identify expression diversifications at the transcription level, which beneficially contribute to infertile men. A lot of studies concentrated on differently expressed gene (DEG) analysis have found a quantity of possible molecular goals and diagnostic biomarkers for infertility at the transcription level. Agnieszka et al. discovers that genes, for example, GGN, GSG1, ADCY10, and GTSF1L are down regulated in human beings with azoospermia. Ribosomal protein S3 (RPS3) is recognized by Zhang et al., RPS5, RPS16, RPS23 and RPS6 were downregulated in teratozoospermia [12, 13].
As the study of male infertility is still insufficient, our goal is to analyze whether there are rare potential disease-related genes associated with infertility, and provide clinical evidence [14]. In this study, a comprehensive bioinformatics method was used to study the related genes of spermatogenesis.
Materials and Methods
Fetching testicular tissue microarray data sets from GEO
The data sets were accessed from GEO (http://www.ncbi.nlm.nih.gov/geo/) in the National Center for Biotechnology Information Database (NCBI) utilizing the accession numbers GSE45885, GSE45887, and GSE9210. Data sets GSE45885 was associated by azoospermia and GSE45887 were presented by Agnieszka Malcher and based on the GPL6244 platform ([HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version, Affymetrix, Inc., USA]). The information of testicular samples in GSE45887 and GSE45885 was accessed from the issued literature (Table 1). The research procedure appeared in Figure 1.
Table 1. Characteristics of GEO sample.
| GSE NO. | Patients | GPL NO. | Experiment type | Organism | Title | Description |
| GSE45885 | 31 | GPL6244 | Expression profiling by array | homo sapiens | Potential biomarkers of non-obstructive azoospermia identified in microarray gene expression analysis | Control group:4;
NOA group:27;
Age:28–54 (yrs). |
| GSE45887 | 20 | GPL6244 | Expression profiling by array | homo sapiens | The gene expression analysis of paracrine/autocrine factors in patients with spermatogenetic failure compared to normal spermatogenesis | Control group:4;
NOA group:16;
Age:28–54 (yrs). |
| GSE9210 | 58 | GPL887 | Expression profiling by array | homo sapiens | A testicular gene expression profile for NOA patients, and ART3 as a genetic susceptibility gene for NOA | 47 non-obstructive azoospermia (NOA) and 11 obstructive azoospermia (OA) patients |
| GSE6872 | 21 | GPL570 | Expression profiling by array | homo sapiens (semen) | Spermatozoal RNA Profiles | Control group:13;
teratozoospermia group:8. |
| GSE108886 | 12 | GPL10558 | Expression profiling by array | homo sapiens (testis) | Spermatogenomics: correlating testicular gene expression to human male infertility | Control group:1;
NOA group:8;
OA group:3. |
| GSE145467 | 20 | GPL4133 | Expression profiling by array | homo sapiens (testis) | Transcriptome changes in patients with severely impaired spermatogenesis | 10 non-obstructive azoospermia (NOA) and 10 obstructive azoospermia (OA) patients |
| GSE4797 | 28 | GPL2891 | Expression profiling by array | homo sapiens | Microarray analysis of human spermatogenic dysfunction | full spermatogenesis (Johnsen Score 10, 12 samples), arrest at the spermatid stage (Johnsen Score 8, 6 samples), arrest at spermatocyte stage (Johnsen Score 5, 5 samples) and Sertoli-cell-only syndrome (Johnsen Score 2, 5 samples). |
| Abbreviations: NOA: non-obstructive azoospermia; OA: obstructive azoospermia. |
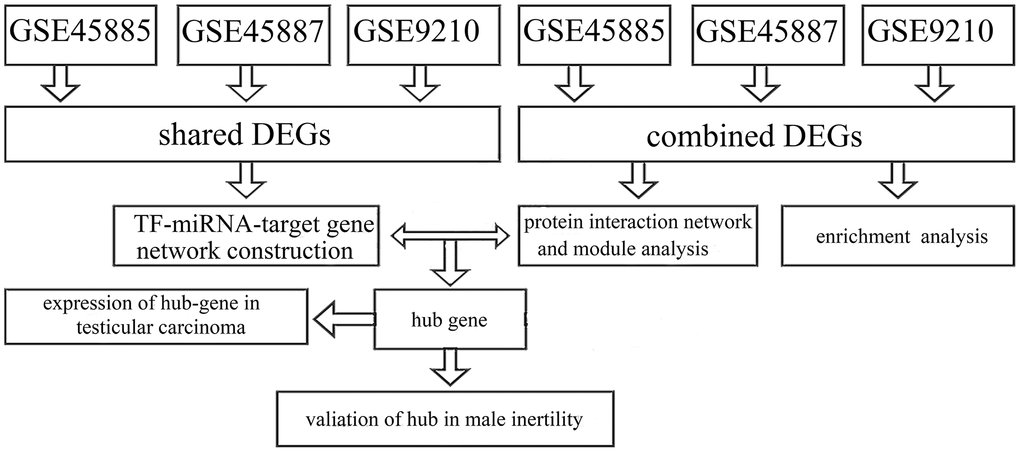
Figure 1. The study procedure.
We fetched gene expression profile (level 3 information) for Level 3 for Testis carcinoma patients from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/).
Microarray data preprocessing
In this study, the original CEL profiles were imported into R (version 3.5, https://www.r-project.org/), and the profiles background was corrected and normalized using the Affy R-package (Bioconductor version 3.6). Affybatch's mas5calls method returns expression sets corresponding to specific genes through multiple probes.
Differentially expressed gene selection
DEGs were used the limma package (version 3.6). |log2 fold change|>1 and P < 0.05 were set at the cutoff values.
Functional annotation and pathway analysis of DEGs
Go analysis is composed of molecular function (MF) and cellular compartment (CC) and biological process (BP) are usual processes for large-scale genomic data's functional annotation. To learn more the mechanism of DEGs that be involved in infertility, we used clusterprofier R-package to analyze the enrichment of KEGG pathways and GO (version 3.16). In these analyses, P < 0.05 was considered statistically significant.
Protein interaction network and module analyses
The STRING database (http://string-db.org), A protein-protein interaction network that was composed of up-regulated and down-regulated DEGs was built, with a cutoff score more than 0.4. Using the clusterone add-in of Cytoscape v3.6.1 to pick the significant modules from the PPI network (https://cytoscape.org/) with P < 0.01 showed statistical importance. The K-core analysis and degree/betweenness/closeness centrality were executed by two add-ins CentiScaPe and Molecular Complex Detection (MCODE) in Cytoscape to illuminate the modules and most significant nodes in the network.
TF-miRNA-target gene network construction
Interactions between differentially expressed miRNAs and differentially expressed mRNAs and expressed miRNAs were forecast utilizing miRWalk 3.0 (http://mirwalk.umm.uni-heidelberg.de/), and a mark 0.95 was regarded to be the cutoff principle for the estimate analysis in miRWalk. The target mRNAs that was involved in all of these databases were only picked for the further analysis. The interaction between mRNA and TF was predicted by using miRWalk 3.0 and the mark 0.4 was considered as cutoff standard for the estimate analysis in the experimental module of LncBase. After the predicted marks were intersected with DEGs, miRNAs, TFs and mRNAs were picked for further analysis. Cytoscape software (version 3.6.1) was used to visualize the regulatory network.
Validation and expression of hub-gene in male infertility and testicular carcinoma
At last, the expression levels of hub genes showed in GEPIA (based on TCGA data) (http://gepia.cancer-pku.cn). Next, validation of hub-gene in male infertility by GEO database (GSE4797, 145467, 108886, 6872) (Table 1). P < 0.05 was viewed to show a statistically important difference in these analyses.
Results
Analysis of DEGs
The expression description information was pre-processed and then analyzed with the Affy package in R language. The entire gene expression was examined. The RNA expression levels are revealed in Figure 2. Differences were revealed by the hierarchical cluster analysis in distribution between normal samples and azoospermic. The consequences showed that grouping was reasonable, and further analysis was undergone by the data with success. Microarray data from the normal samples were compared with those from the azoospermic samples and a sum of 1396 DEGs were discovered.
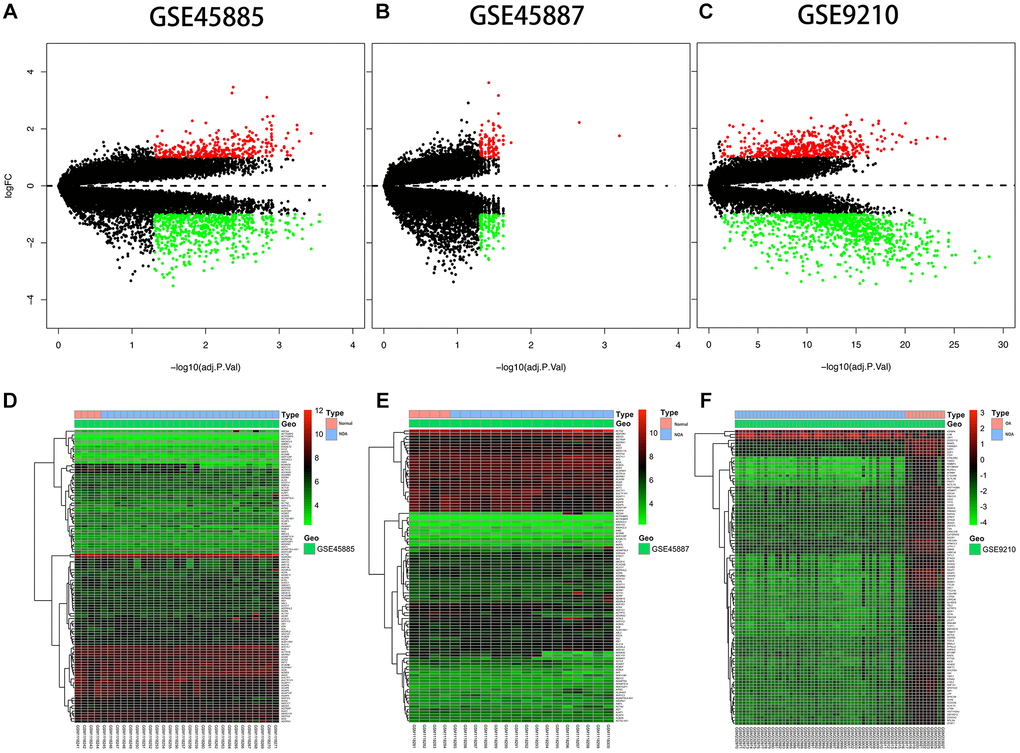
Figure 2. Differential expression analysis. (A, B, C) Volcano plot of DEGs. The y-axis is logFC and the x-axis represents -log10 (adjusted P-value). The red dots represent the DEGs upregulated and the green dots represent the DEGs downregulated while the black dots represent genes that were not differentially expressed. DEGs, differentially expressed genes; FC, fold change. (D, E, F) Heat map presenting the expression pattern across different samples. The horizontal axis represents sample names. The left vertical axis presented clusters of DEGs, and the top horizontal axis presents clusters of samples. Red represents upregulated genes and green represents downregulated genes.
Functional annotation and pathway analysis of DEGs
A total of 1396 genes were identified by enrichment analysis, with P (by less than 0.05) statistical significance used to be determined. Figure 3A–3C displayed that GO-MF, CC, BP. The top 10 GO terms that are enriched through up and downregulated genes were primarily enriched in “tubulin binding”, “cyclin-dependent protein serine/threonine kinase regulator activity”, “steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor”, “motor activity”, “protein binding involved in protein folding”, “microtubule binding”, “glutathione transferase activity”, “dynein heavy chain binding”, “steroid dehydrogenase activity”, “extracellular matrix structural constituent” (Figure 3D). The KEGG pathways of DEGs (Figure 3E) were primarily enriched in “Oocyte meiosis”, “Human T-cell leukemia virus 1 infection”, “cell cycle”, “progesterone-mediated oocyte maturation”, “glucagon signaling pathway”, “foxo signaling pathway”, “staphylococcus aureus infection”, “aldosterone synthesis and secretion”, “toxoplasmosis”, “carbon metabolism”. Figure 3F demonstrated 27 DEGs co-expression in three GEO data sets.
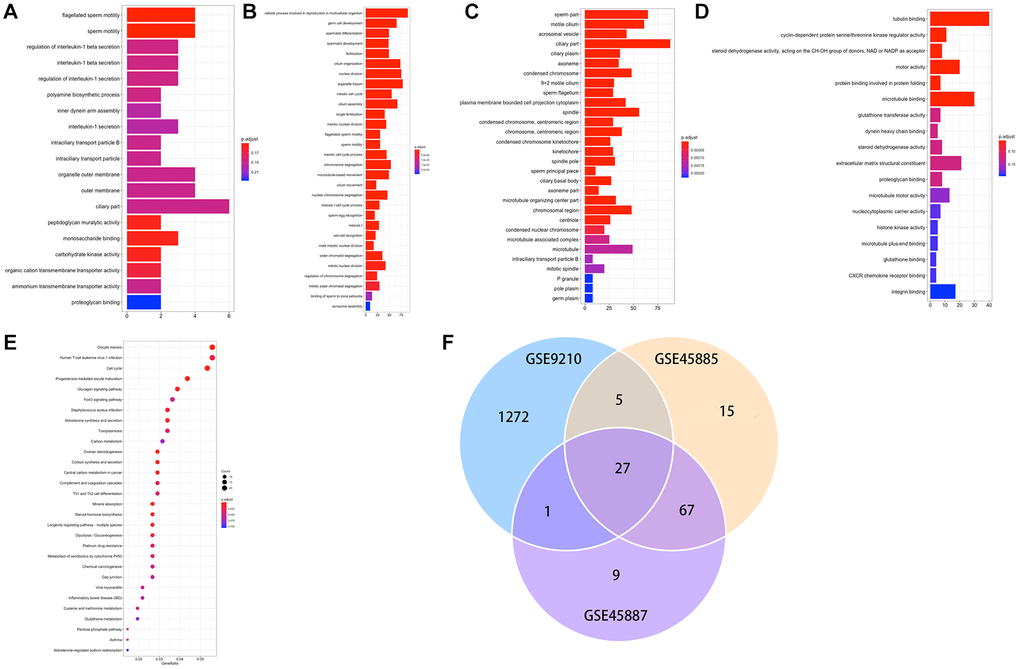
Figure 3. (A–E) GO and KEGG results of DEGs. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; (F) Venn plot of candidate hub genes commonly owned in GSE45885, GSE45887 and GSE9210.
Protein interaction network and module analysis
To investigate shared genes' relationship between modules, PPI networks were built by using Cytoscape software that was based on the STRING database. Moreover, the k-core analysis was executed to discover the hub genes and cardinal clusters of PPI networks. By Cytoscape-MCODE analysis, parameters are set as follows: Degree Cutoff: 5, Node Score Cutoff: 0.2, K-Core: 5. 10 clusters of 338 spermatogenesis-associated genes (Figure 4) were identified by us. The hub genes in cluster 1 (Figure 4) that exhibited the highest scores (52.59) were TOP2A, CDT1, KPNA2, TACC3, TEKT2,NUF2, ATAD2, PBK, DLGAP5, TYMS, KIF18A, PTTG1, SPAG5, CDCA8, AURKA, EZH2, CCNB2, EXO1, NCAPG, SMC2, BUB1, KIF15, CDK1, CDC45, ZWILCH, KNTC1, NEK2, KIF20A, MCM4, MAD2L1, CDCA5, CDC20, CCNB1, CENPK, CKS2, OIP5, HMMR, PLK4, ASPM, CDKN3, CEP55, RAD54L, TTK, KIF2C, CENPF, CDCA2, SKA3, SGOL2, RAD51, SPC25, RFC4, MND1, CENPM, CENPU, CASC5, BIRC5, CDC25C, GMNN, RACGAP1, ANLN, UHRF1.
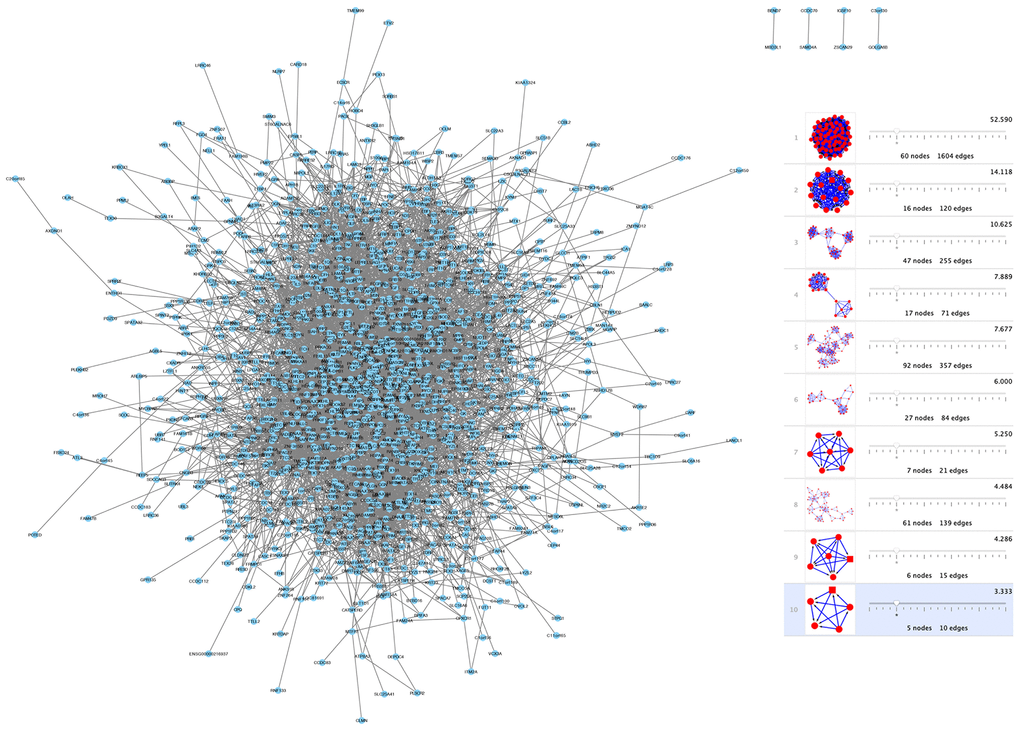
Figure 4. Network and module analysis of DEGs. (left) PPI network of DEGs obtained from the STRING database. (right) 10 clusters identified through Cytoscape-MCODE analysis. Abbreviations: DEGs: differentially expressed genes; PPI: protein-protein interaction.
TF-miRNA-target gene network construction
A total of 42 miRNAs could bind to shared genes predicted by miRWalk. Four genes did not have any binding miRNA. The regulatory network of TF-miRNA-target gene was established, involving 18 TFs and 23 hub genes in Figure 5, such as KIF2C-miRNAs (has-miR-3154, 6075, 6760-5p, 1251-5p, 186-sp)-TFs (EP300, SP1).
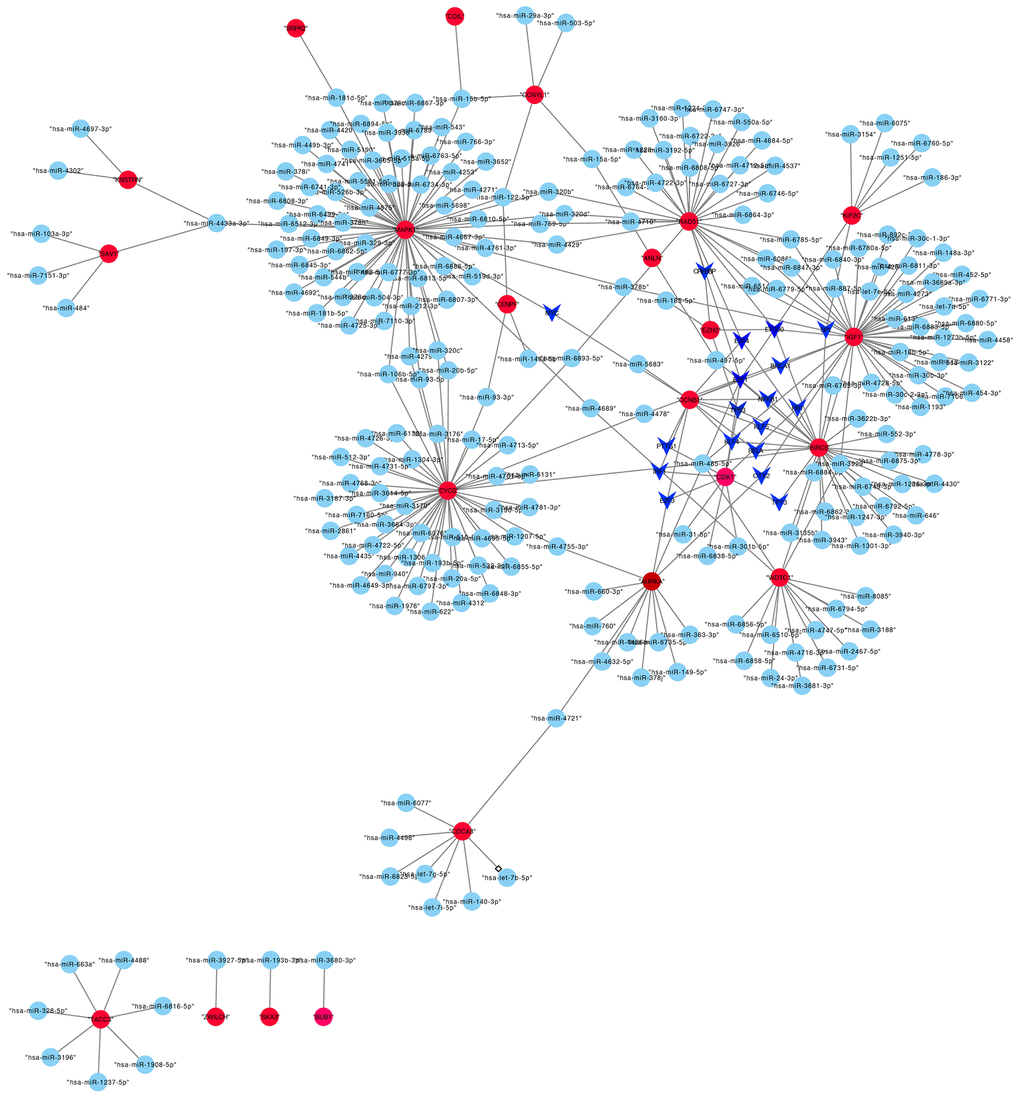
Figure 5. The transcriptional regulatory network of hub genes, miRNAs, and TFs. Abbreviations: miRNAs, microRNAs; TFs, transcription factors.
Expression of hub-gene in testicular carcinoma
A total of 12 genes (ANLN, CCNB1, CENPF, COIL, CYCS, KIF2C, KNSTRN, LELP1, OAZ3, SRPK2, TEKT2, WDTC1) were identified as hub genes. We used TCGA data of testis cancer to validate the hub gene expression with the online tool of GEPIA. All of the hub genes are expressed differently in normal and cancer tissues of testis cancer by the criterion of |logFC|>1 and p < .01 (Figure 6).
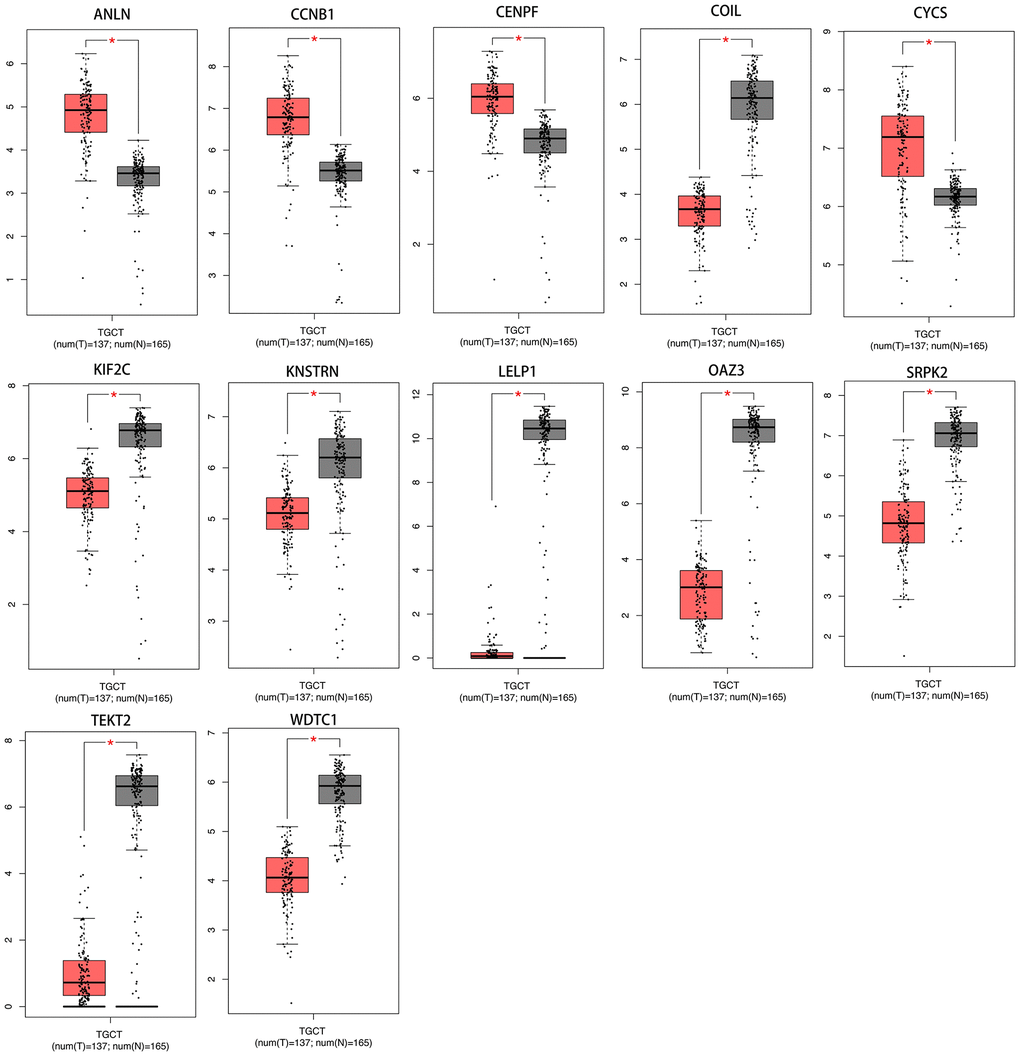
Figure 6. The transcriptional differences of hub gene levels between colon carcinoma tissues and the para-cancer tissues in TCGA. TCGA, The Cancer Genome Atlas (*p < .001).
Validation of hub-gene in male infertility
Through validation of hub-gene (A total of 12 genes were identified as hub genes in testicular carcinoma) in male infertility by GSE4797 set, we found expression of KIF2C between more than or equal to John score 5 and less than 5 existed significant difference. The expression of TEKT2 decreased with reduction of John score in the Figure 7. The Figure 8 showed the expression of KIF2C and TEKT2 both decreased in three datasets from GEO database.
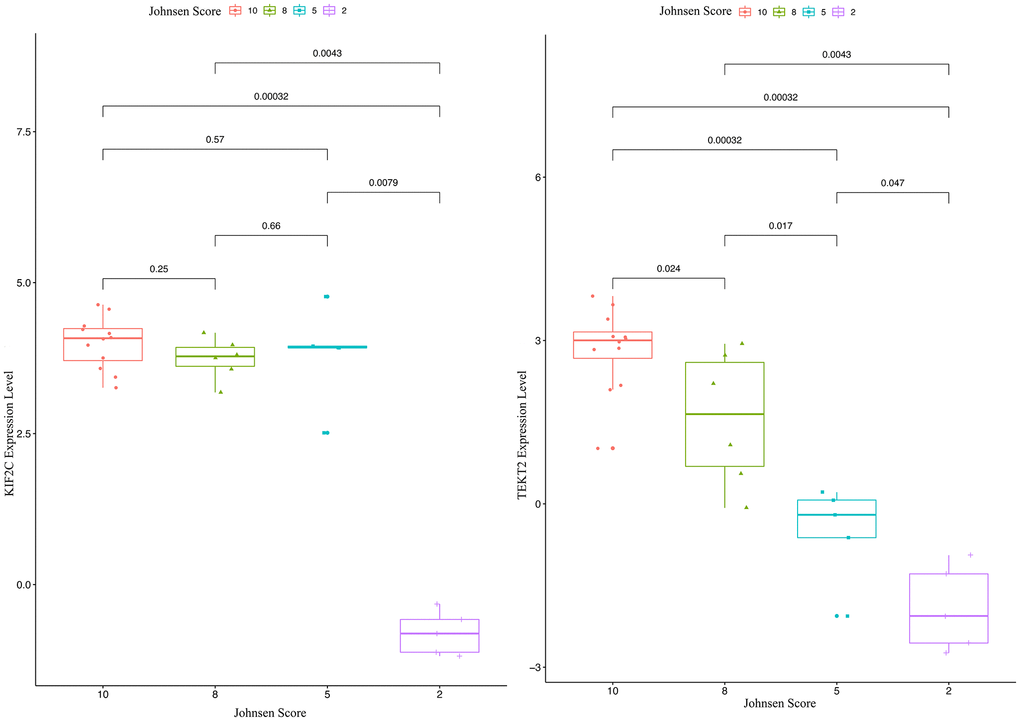
Figure 7. The validation of KIF2C and TEKT2 in GSE4797 associated with male infertility.
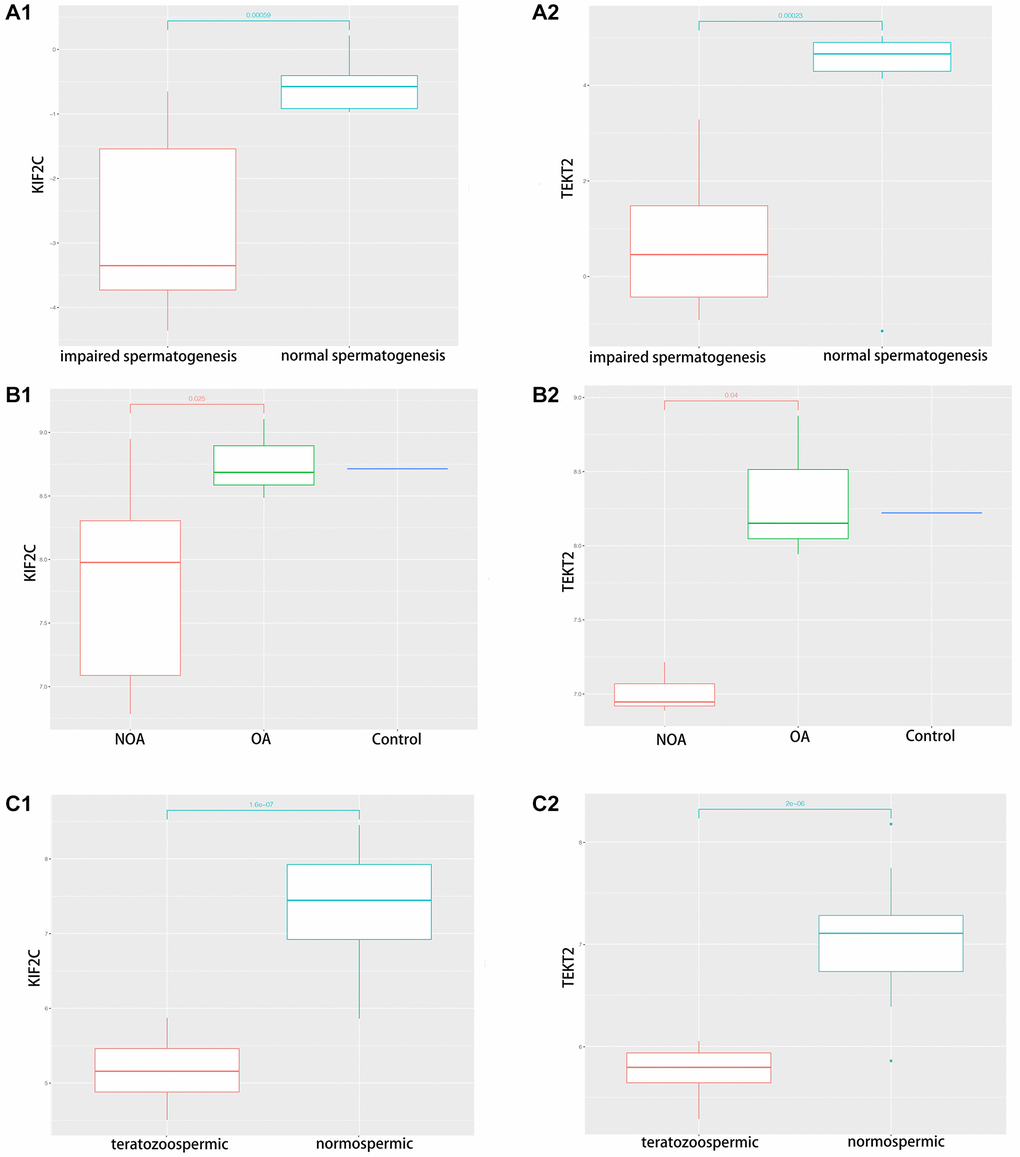
Figure 8. The validation of KIF2C and TEKT2 in GEO database associated with spermatogenesis. (A) GSE145467; (B) GSE108886; (C) GSE6872.
Furthermore, we found KIF2C might act in infertility and testis cancer through DEG, functional annotation and pathway, protein interaction network and module analysis.
Discussion
With the development of science and technology, the molecular mechanism of azoospermia has aroused great interest. The study of azoospermia depends on human studies, animal models, organ culture models and cell lines [9, 15]. It has been confirmed that the number of genes and proteins related to male infertility has increased [9, 16]. However, how these biological processes are regulated at the molecular level remains to be elucidated. Therefore, it is necessary to further study the pathogenesis of azoospermia at the molecular level. In this study, we identified the genes associated with spermatogenesis and systematically constructed a comprehensive framework of genes and miRNAs.
In the present study, we sought to identify associated genes by reanalyzing separate studies from GEO datasets (GSE45885, GSE45887, and GSE9210) and validation dataset (GSE4797, 145467, 108886 and 6872) [13, 17, 18]. The results showed that several shared genes associated with spermatogenesis. Finally, we effectively screen out two genes (KIF2C and TEKT2) for validation by GSE4797, 145467, 108886 and 6872. Among these two genes, KIF2C and TEKT2 down-regulated in sperm abnormality. The network of TF-miRNA-target gene was established, we found KIF2C-miRNAs (has-miR-3154, 6075, 6760-5p, 1251-5p, 186-sp)-TFs (EP300, SP1) might work in spermatogenesis. Interestingly, the relative expression levels of KIF2C and TEKT2 had a negative correlation with Johnsen score, which showed potential role of spermatogenesis.
KIF2C (also known as the mitotic centromere-associated kinesin, MACK) is a member of the kinesin-13 family of microtubule (MT)-depolymerizing kinesins, which is critical in the regulation of microtubule dynamics. During cell division, KIF2C inhibits the wrong connection between MT and chromosome [19, 20]. The function of KIF2C in interphase cells is not obvious, although its main localization in nucleus suggests that KIF2C may work in nuclear processes. KIF2C promotes the formation of DNA damage foci, which may involve the migration and aggregation of DSBs (DNA Double Strand Break) [21–25]. We found KIF2C might work in the testis cancer and spermatogenesis.
Tektins (TEKTs), the proteins of the microtubules in Cilia, Flagella, Basal bodies and centrioles [26–28], have been found in various animals, including Filariae, including silk-worms [29], mice [26, 30] and humans [31, 32]. They were originally isolated from sea urchins and are a group of proteins: TEKT-A, -B and -C [33, 34]. On the other hand, five types of TEKTs have been identified in mammals. TEKT2, which is similar to Tektin-t, locates in the main part of human spermatozoa but no immune signal was detected in the middle or at the end of the human sperm. Tektin2, a membrane protein, is responsible for sperm flagellum movement. Previous studies show that CatSper and tektin are associated with male infertility because they play an important role in sperm motility [35]. Tektin2 is essential for the integrity of motilin arm in sperm flagellum. Lack of tektin2 can lead to impaired sperm motility and male infertility [36]. The low expression of tektin2 mRNA was observed in frozen spermatozoa, suggesting that the decrease of sperm motility after cryopreservation may be due to the transcriptional damage of some sperm motility related genes [37].
The miRNAs work in infertility. In 2009, for the first time, expression of miRNAs in a testicular sample of NOA patients compared to fertile control samples evaluated by microarray technology, identified 19 upregulated and 154 downregulated miRNAs [38]. Hsa-miR-141, hsa-miR-429, hsa-miR-7-1- 3p, hsa-miR-34b, hsa-miR-34c-5p, hsa-miR-122 expression levels were different in azoospermia [39, 40]. Through luciferase experiments, miR-525-3p which targets SEMG1 gene and hsa-miR-210 which targets insulin-like growth factor II (IGF2) [41, 42]. The lower expression of hsa-miR-188-3p results in higher expression of MLH1 gene in azoospermia patients and leads to apoptosis in spermatozoa [43].
Functional classification of the miRNA/mRNA pairs using bioinformatics tools indicated that they play a role in spermatogenesis, cell meiosis, cell cycle. We found KIF2C-miRNAs (has-miR-3154, 6075, 6760-5p, 1251-5p, 186-sp)-TFs (EP300, SP1) might work in spermatozoa of infertile men.
Our study also has some limitations. First, more samples could be included in this study and we assessed our results based on published observations. Further in vitro and/or in vivo experiments would need to be carried out to test reliability of our results. This might reduce the error caused by individual differences of patients.
Conclusions
We applied DEG analysis to identify genes associated with azoospermia in this study. Then, through a system biology framework for a comprehensive and systematic biological function- and network-based analysis of azoospermia, we found 27 hub genes and test on the expression of hub-gene in testicular carcinoma (found 12 hub genes were different in testicular carcinoma). Furthermore, we made the validation of hub-gene (A total of 12 genes were identified as hub genes in testicular carcinoma) in male infertility by GSE4797, 145467, 108886 and 6872 and found TEKT2 and KIF2C might work in infertility. The network of TF-miRNA-target gene was established and we found KIF2C-miRNAs (has-miR-3154, 6075, 6760-5p, 1251-5p,186-sp)-TFs (EP300, SP1) might work in spermatozoa of infertile men. Our study might help to improve our understanding of the mechanisms in azoospermia and provide diagnostic biomarkers and therapeutics targets.
Abbreviations
GEO: Gene Expression Omnibus database;
TCGA: The cancer genome atlas;
DEG: differently expressed gene;
GGN: gametogenetin;
GSG1: germ cell associated 1;
ADCY10: adenylate cyclase 10;
GTSF1L: gametocyte-specific factor 1 like;
NCBI: National Center for Biotechnology Information Database;
BP: biological process;
CC: cellular compartment;
MF: molecular function;
GEPIA: Gene Expression Profiling Interactive Analysis;
FC: fold change;
GO: Gene Ontology;
KEGG: Kyoto Encyclopedia of Genes and Genomes;
TF: Transcript factor;
KIF2C: Kinesin Family Member 2C;
TEKT2: Tektin 2;
MACK: mitotic centromere-associated kinesin;
DSBs: DNA Double Strand Break;
NOA: Non-obstructive azoospermia;
MT: microtubule;
IGF2: insulin-like growth factor II.
Author Contributions
HMC and ZW conceived the study, performed bioinformatic analysis, analyzed and interpreted the data and wrote the manuscript. FW and ZYL contributed to resources, revised the manuscript. XFL and JQH conceived the study, supervised the study, analyzed and interpreted the data, gave the final approval of the manuscript.
Acknowledgments
We are grateful for our colleague’s help.
Conflicts of Interest
The authors declare no conflicts of interest related to this study.
Funding
This work was supported by Bengbu Medical College Innovation project for graduate student (NO. BYYCX20089).
References
-
1.
Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID, Simpson JL, van der Poel S. The International Glossary on Infertility and Fertility Care, 2017. Fertil Steril. 2017; 108:393–406. https://doi.org/10.1016/j.fertnstert.2017.06.005 [PubMed]
-
2.
Vander Borght M, Wyns C. Fertility and infertility: Definition and epidemiology. Clin Biochem. 2018; 62:2–10. https://doi.org/10.1016/j.clinbiochem.2018.03.012 [PubMed]
-
3.
Barak S, Gordon Baker HW. Chapter 141 - Clinical management of male infertility, ed(s): Jameson JL, de Groot LJ, de Kretser DM, Giudice LC, Grossman AB, Melmed S, Potts JT, Weir GC. Endocrinology: Adult and Pediatric (7th ed), Saunders WB, 2016, pp 2431–55.e4. https://doi.org/10.1016/B978-0-323-18907-1.00141-4 [PubMed]
-
4.
ESHRE Capri Workshop Group. A prognosis-based approach to infertility: understanding the role of time. Hum Reprod. 2017; 32:1556–59. https://doi.org/10.1093/humrep/dex214 [PubMed]
-
5.
Briceag I, Costache A, Purcarea VL, Cergan R, Dumitru M, Briceag I, Sajin M, Ispas AT. Fallopian tubes--literature review of anatomy and etiology in female infertility. J Med Life. 2015; 8:129–31. [PubMed]
-
6.
Kumar N, Singh AK. Trends of male factor infertility, an important cause of infertility: A review of literature. J Hum Reprod Sci. 2015; 8:191–96. https://doi.org/10.4103/0974-1208.170370 [PubMed]
-
7.
Masoumi SZ, Parsa P, Darvish N, Mokhtari S, Yavangi M, Roshanaei G. An epidemiologic survey on the causes of infertility in patients referred to infertility center in Fatemieh Hospital in Hamadan. Iran J Reprod Med. 2015; 13:513–16. [PubMed]
-
8.
Agarwal A, Mulgund A, Hamada A, Chyatte MR. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015; 13:37. https://doi.org/10.1186/s12958-015-0032-1 [PubMed]
-
9.
Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol. 2018; 15:369–84. https://doi.org/10.1038/s41585-018-0003-3 [PubMed]
-
10.
Tournaye H, Krausz C, Oates RD. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017; 5:544–53. https://doi.org/10.1016/S2213-8587(16)30040-7 [PubMed]
-
11.
Krausz C. Male infertility: pathogenesis and clinical diagnosis. Best Pract Res Clin Endocrinol Metab. 2011; 25:271–85. https://doi.org/10.1016/j.beem.2010.08.006 [PubMed]
-
12.
Zhang T, Wu J, Liao C, Ni Z, Zheng J, Yu F. System analysis of teratozoospermia mRNA profile based on integrated bioinformatics tools. Mol Med Rep. 2018; 18:1297–304. https://doi.org/10.3892/mmr.2018.9112 [PubMed]
-
13.
Malcher A, Rozwadowska N, Stokowy T, Kolanowski T, Jedrzejczak P, Zietkowiak W, Kurpisz M. Potential biomarkers of nonobstructive azoospermia identified in microarray gene expression analysis. Fertil Steril. 2013; 100:1686–94.E7. https://doi.org/10.1016/j.fertnstert.2013.07.1999 [PubMed]
-
14.
Barratt CLR, Björndahl L, De Jonge CJ, Lamb DJ, Osorio Martini F, McLachlan R, Oates RD, van der Poel S, St John B, Sigman M, Sokol R, Tournaye H. The diagnosis of male infertility: an analysis of the evidence to support the development of global WHO guidance-challenges and future research opportunities. Hum Reprod Update. 2017; 23:660–80. https://doi.org/10.1093/humupd/dmx021 [PubMed]
-
15.
Babakhanzadeh E, Nazari M, Ghasemifar S, Khodadadian A. Some of the Factors Involved in Male Infertility: A Prospective Review. Int J Gen Med. 2020; 13:29–41. https://doi.org/10.2147/ijgm.s241099 [PubMed]
-
16.
Araujo TF, Friedrich C, Grangeiro CHP, Martelli LR, Grzesiuk JD, Emich J, Wyrwoll MJ, Kliesch S, Simões AL, Tüttelmann F. Sequence analysis of 37 candidate genes for male infertility: challenges in variant assessment and validating genes. Andrology. 2020; 8:434–41. https://doi.org/10.1111/andr.12704 [PubMed]
-
17.
Malcher A, Rozwadowska N, Stokowy T, Jedrzejczak P, Zietkowiak W, Kurpisz M. The gene expression analysis of paracrine/autocrine factors in patients with spermatogenetic failure compared with normal spermatogenesis. Am J Reprod Immunol. 2013; 70:522–28. https://doi.org/10.1111/aji.12149 [PubMed]
-
18.
Feig C, Kirchhoff C, Ivell R, Naether O, Schulze W, Spiess AN. A new paradigm for profiling testicular gene expression during normal and disturbed human spermatogenesis. Mol Hum Reprod. 2007; 13:33–43. https://doi.org/10.1093/molehr/gal097 [PubMed]
-
19.
Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007; 18:2970–79. https://doi.org/10.1091/mbc.e07-02-0110 [PubMed]
-
20.
Sanhaji M, Friel CT, Wordeman L, Louwen F, Yuan J. Mitotic centromere-associated kinesin (MCAK): a potential cancer drug target. Oncotarget. 2011; 2:935–47. https://doi.org/10.18632/oncotarget.416 [PubMed]
-
21.
Asaithamby A, Chen DJ. Mechanism of cluster DNA damage repair in response to high-atomic number and energy particles radiation. Mutat Res. 2011; 711:87–99. https://doi.org/10.1016/j.mrfmmm.2010.11.002 [PubMed]
-
22.
Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004; 303:92–95. https://doi.org/10.1126/science.1088845 [PubMed]
-
23.
Aymard F, Aguirrebengoa M, Guillou E, Javierre BM, Bugler B, Arnould C, Rocher V, Iacovoni JS, Biernacka A, Skrzypczak M, Ginalski K, Rowicka M, Fraser P, Legube G. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat Struct Mol Biol. 2017; 24:353–61. https://doi.org/10.1038/nsmb.3387 [PubMed]
-
24.
Neumaier T, Swenson J, Pham C, Polyzos A, Lo AT, Yang P, Dyball J, Asaithamby A, Chen DJ, Bissell MJ, Thalhammer S, Costes SV. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc Natl Acad Sci U S A. 2012; 109:443–48. https://doi.org/10.1073/pnas.1117849108 [PubMed]
-
25.
Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013; 341:660–64. https://doi.org/10.1126/science.1237150 [PubMed]
-
26.
Norrander J, Larsson M, Ståhl S, Höög C, Linck R. Expression of ciliary tektins in brain and sensory development. J Neurosci. 1998; 18:8912–18. https://doi.org/10.1523/JNEUROSCI.18-21-08912.1998 [PubMed]
-
27.
Steffen W, Linck RW. Evidence for tektins in centrioles and axonemal microtubules. Proc Natl Acad Sci U S A. 1988; 85:2643–47. https://doi.org/10.1073/pnas.85.8.2643 [PubMed]
-
28.
Larsson M, Norrander J, Gräslund S, Brundell E, Linck R, Ståhl S, Höög C. The spatial and temporal expression of Tekt1, a mouse tektin C homologue, during spermatogenesis suggest that it is involved in the development of the sperm tail basal body and axoneme. Eur J Cell Biol. 2000; 79:718–25. https://doi.org/10.1078/0171-9335-00097 [PubMed]
-
29.
Ota A, Kusakabe T, Sugimoto Y, Takahashi M, Nakajima Y, Kawaguchi Y, Koga K. Cloning and characterization of testis-specific tektin in Bombyx mori. Comp Biochem Physiol B Biochem Mol Biol. 2002; 133:371–82. https://doi.org/10.1016/s1096-4959(02)00153-7 [PubMed]
-
30.
Iguchi N, Tanaka H, Fujii T, Tamura K, Kaneko Y, Nojima H, Nishimune Y. Molecular cloning of haploid germ cell-specific tektin cDNA and analysis of the protein in mouse testis. FEBS Lett. 1999; 456:315–21. https://doi.org/10.1016/s0014-5793(99)00967-9 [PubMed]
-
31.
Xu M, Zhou Z, Cheng C, Zhao W, Tang R, Huang Y, Wang W, Xu J, Zeng L, Xie Y, Mao Y. Cloning and characterization of a novel human TEKTIN1 gene. Int J Biochem Cell Biol. 2001; 33:1172–82. https://doi.org/10.1016/s1357-2725(01)00089-9 [PubMed]
-
32.
Wolkowicz MJ, Naaby-Hansen S, Gamble AR, Reddi PP, Flickinger CJ, Herr JC. Tektin B1 demonstrates flagellar localization in human sperm. Biol Reprod. 2002; 66:241–50. https://doi.org/10.1095/biolreprod66.1.241 [PubMed]
-
33.
Linck RW, Albertini DF, Kenney DM, Langevin GL. Tektin filaments: chemically unique filaments of sperm flagellar microtubules. Prog Clin Biol Res. 1982; 80:127–32. https://doi.org/10.1002/cm.970020724 [PubMed]
-
34.
Norrander JM, Perrone CA, Amos LA, Linck RW. Structural comparison of tektins and evidence for their determination of complex spacings in flagellar microtubules. J Mol Biol. 1996; 257:385–97. https://doi.org/10.1006/jmbi.1996.0170 [PubMed]
-
35.
Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, Krapivinsky L, Kirichok Y, Ramsey IS, Quill TA, Clapham DE. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc Natl Acad Sci U S A. 2007; 104:1219–23. https://doi.org/10.1073/pnas.0610286104 [PubMed]
-
36.
Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, Maekawa M, Nishimune Y. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol Cell Biol. 2004; 24:7958–64. https://doi.org/10.1128/MCB.24.18.7958-7964.2004 [PubMed]
-
37.
Valcarce DG, Cartón-García F, Herráez MP, Robles V. Effect of cryopreservation on human sperm messenger RNAs crucial for fertilization and early embryo development. Cryobiology. 2013; 67:84–90. https://doi.org/10.1016/j.cryobiol.2013.05.007 [PubMed]
-
38.
Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang C, Li X, Sun F. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2009; 7:13. https://doi.org/10.1186/1477-7827-7-13 [PubMed]
-
39.
Wu W, Qin Y, Li Z, Dong J, Dai J, Lu C, Guo X, Zhao Y, Zhu Y, Zhang W, Hang B, Sha J, Shen H, et al. Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: significant up-regulation of miR-141, miR-429 and miR-7-1-3p. Hum Reprod. 2013; 28:1827–36. https://doi.org/10.1093/humrep/det099 [PubMed]
-
40.
Abu-Halima M, Hammadeh M, Backes C, Fischer U, Leidinger P, Lubbad AM, Keller A, Meese E. Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertil Steril. 2014; 102:989–97.e1. https://doi.org/10.1016/j.fertnstert.2014.07.001 [PubMed]
-
41.
Zhou QZ, Guo XB, Zhang WS, Zhou JH, Yang C, Bian J, Chen MK, Guo WB, Wang P, Qi T, Wang CY, Yang JK, Liu CD. Expressions of miR-525-3p and its target gene SEMG1 in the spermatozoa of patients with asthenozoospermia. Andrology. 2019; 7:220–27. https://doi.org/10.1111/andr.12573 [PubMed]
-
42.
Tang D, Huang Y, Liu W, Zhang X. Up-Regulation of microRNA-210 is Associated with Spermatogenesis by Targeting IGF2 in Male Infertility. Med Sci Monit. 2016; 22:2905–10. https://doi.org/10.12659/msm.897340 [PubMed]
-
43.
Song WY, Meng H, Wang XG, Jin HX, Yao GD, Shi SL, Wu L, Zhang XY, Sun YP. Reduced microRNA-188-3p expression contributes to apoptosis of spermatogenic cells in patients with azoospermia. Cell Prolif. 2017; 50:e12297. https://doi.org/10.1111/cpr.12297 [PubMed]