Sevoflurane upregulates neuron death process-related Ddit4 expression by NMDAR in the hippocampus
Abstract
Postoperative cognitive dysfunction (POCD) is a serious and common complication induced by anesthesia and surgery. Neuronal apoptosis induced by general anesthetic neurotoxicity is a high-risk factor. However, a comprehensive analysis of general anesthesia-regulated gene expression patterns and further research on molecular mechanisms are lacking. Here, we performed bioinformatics analysis of gene expression in the hippocampus of aged rats that received sevoflurane anesthesia in GSE139220 from the GEO database, found a total of 226 differentially expressed genes (DEGs) and investigated hub genes according to the number of biological processes in which the genes were enriched and performed screening by 12 algorithms with cytoHubba in Cytoscape. Among the screened hub genes, Agt, Cdkn1a, Ddit4, and Rhob are related to the neuronal death process. We further confirmed that these genes, especially Ddit4, were upregulated in the hippocampus of aged mice that received sevoflurane anesthesia. NMDAR, the core target receptor of sevoflurane, rather than GABAAR, mediates the sevoflurane regulation of DDIT4 expression. Our study screened sevoflurane-regulated DEGs and focused on the neuronal death process to reveal DDIT4 as a potential target mediated by NMDAR, which may provide a new target for the treatment of sevoflurane neurotoxicity.
Introduction
More than 300 million operations are performed worldwide each year, and a continued increase is observed in all economic environments [1]. Postoperative cognitive dysfunction (POCD) is a common cognitive impairment in patients during the perioperative period and is mostly found in elderly patients, with a reported incidence ranging from 15% to 60% [2, 3]. POCD is mainly characterized by progressive postoperative memory impairment, cognitive decline, and executive dysfunction. In addition, the effects of POCD may not be temporary and can lead to neurological dysfunction years after surgery [4, 5], which is associated with an increased risk of life-threatening illness and death. Neuronal apoptosis, a high-risk factor inducing POCD, leads to decreased neurogenesis, impaired synaptic plasticity, neuroinflammation, and oxidative stress in POCD patients [6–8]. Sevoflurane can induce POCD-related behaviors in animal, such as mice [9] or rats [10, 11]. However, the neurobiological basis of sevoflurane neurotoxicity remains largely unknown.
General anesthetic neurotoxicity has been extensively examined in recent years [12–14]. An increasing number of studies have shown that inhaled anesthetics may cause neurotoxicity, leading to hippocampal neuronal damage and apoptosis, which result in cognitive dysfunction [15–17]. Sevoflurane, the most commonly used inhalation anesthetic, induces neuronal apoptosis [18–22]. Sevoflurane enhanced the production of lactate in aged marmoset brains [23]. Lactate accumulation can induce neuronal apoptosis or even acidosis in critically ill patients [24–26]. Sevoflurane was shown to activate gamma-aminobutyric acid subtype A receptor (GABAAR) to induce apoptosis of immature dentate granule cells in mice [27]. Apoptosis is regulated by multiple pathways, among which the mechanism of neuronal apoptosis induced by sevoflurane through related signaling pathways has attracted increased attention [28–30]. Sevoflurane inhibits the ERK1/2 signaling pathway by antagonizing the N-methyl-D-aspartate receptor (NMDAR) and upregulates the expression of the apoptotic proteins caspase-3 and Bax in mitochondria, resulting in apoptosis of hippocampal neurons [31]. Additionally, sevoflurane promotes the expression of the apoptotic factor connexin 43 (Cx43) and leads to neuronal apoptosis by activating the JNK/cJun/AP-1 signaling pathway [32]. However, a comprehensive analysis of the differentially expressed genes (DEGs) regulated by sevoflurane and further investigation of the molecular mechanism is lacking.
Transcriptomic analysis has identified comprehensive gene expression patterns to help reveal potential mechanisms for various neurological diseases [33, 34] and has shown that inhaled anesthetic are associated with neurological damage [35]. Here, we applied multistage comprehensive bioinformatics methods to explore the possible pathogenesis of sevoflurane neurotoxicity. We focused on the potential key genes with different expression levels in the hippocampus of aged rats after sevoflurane anesthesia, established the functional annotation of their potential target genes and used gene enrichment analysis to reveal the role of DEGs associated with the cell death process in sevoflurane anesthesia. We further performed an in vivo experiment in aged mice that received sevoflurane exposure to confirm the pattern of upregulation of these genes in the hippocampus and further found that the NMDAR mediates the sevoflurane regulation of DDIT4 expression. This work showed the molecular mechanism of sevoflurane-induced neuronal apoptosis and provided a new potential target for sevoflurane toxicity.
Results
DEG identification
A total of 10032 unique genes were annotated with the SwissProt database. The expression boxplot of all genes for each sample is shown in Figure 1A after normalization with the rma function by using the oligo package. The differential expression analysis identified 194 upregulated and 32 downregulated genes after treatment with 2.5% sevoflurane in 100% oxygen for 4 hours in an anesthetizing chamber with the criterion of a P value less than 0.05. Among all DEGs, 160 DEGs (153 upregulated DEGs and 17 downregulated DEGs) could be annotated in metascape, they were listed in Table 1, and the heatmap of the DEGs between the two groups is displayed in Figure 1B.
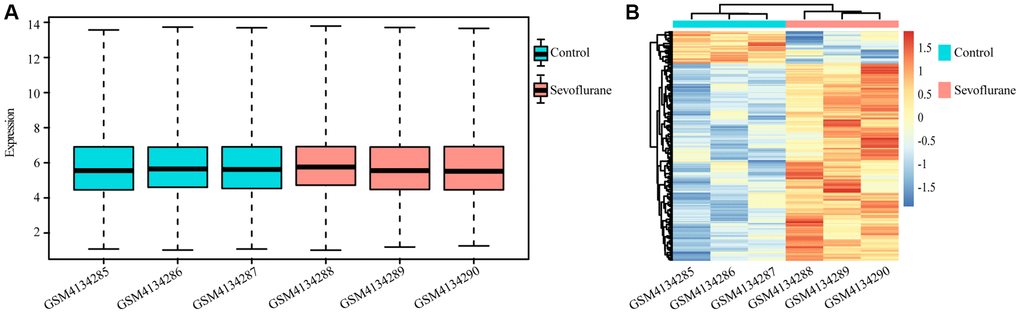
Figure 1. Differentially regulated genes between the control and sevoflurane-treated groups. (A) Boxplot of all genes in each sample. (B) Heatmap of differentially regulated genes in the control and sevoflurane-treated groups, 194 upregulated and 32 downregulated genes after treatment with 2.5% sevoflurane in 100% oxygen for 4 hours. (Green: Control group; Red: sevoflurane-treated group).
Table 1. Differentially regulated genes between the control and sevoflurane-treated groups.
| Differentially regulated genes |
| 153 Upregulated genes in the sevoflurane-treated group. | Slc19a3; Tmem163; Snx9; Ptp4a3; Rabif; Rnf125; Creg1; Dbt; Ccdc115; Bcas2; Abhd15; Lsm5; Olfr1283; Pex10; Olfr1499; Olfr273; Timm29; Lonrf3; Map3k6; Pla2g3; Tfcp2l1; Olfr325; Olfr432; Serpinb3a; Krtap13; Hddc2; BC005624; Olfr362; Hipk2; Slc2a12; Olfr1406; Acer2; Arrdc2; Zfp521; Peli2; Sun2; Mocs1; Parvg; Ctxn2; Slc2a9; Cldn2; Srp9; Ecrg4; Sf3b5; Ikzf2; Ikzf1; Top3b; Mlc1; Fam83d; Trim21; Pdcd5; Mrgprb3; Gm266; Olfr1321; Tnfrsf11a; Dlc1; Lrig3; Sult1d1; Elmo1; Olfr1451; Olfr1299; H1f3; Tceal5; Ddah1; Fxyd1; Gpd1; Smarcd2; Nfe2l2; Pex11a; Ackr3; Agt; C3; Pla2g1b; Gsta3; A2m; Mag; Aldoc; Gpr139; Sparc; Plat; Pdgfra; Ptgds; Tgfbr3; Oprm1; Rpl36; Aqp4; Tmbim6; Mertk; Phactr2; Rhob; Hrk; Pcp4; Timp4; Hand1; Klf9; Cntfr; Afg1l; Adipor2; Fam43a; Rgs16; Ehd2; Spint1; Rassf4; Prr5; Gramd3; Wdr89; Klra8; Lims2; Cnksr3; Rac2; Fkbp14; Ech1; Plcd4; Acsl3; Gbp2; Prlhr; Cdkn1a; Prkg2; Prkch; Psat1; Etfb; Tmem252; Gsto2; Prxl2a; Paqr8; Rab31; Usp54; Clic1; Bspry; Cnppd1; Slc10a6; Slc3a2; Zfp24; Slc29a3; Rcan2; Ddit4; Dhrs4; Cygb; Igsf1; Plce1; Tsc22d3; Zfp422; Slc38a2; Plcb4; Slc12a2; Pip4k2a; Apln; Olig1; Timm8a1; Ppm1f; Rapgef3 |
| 17 Downregulated genes in the sevoflurane-treated group. | Olfr1461; Flrt2; Kif21b; Olfr1474; Kcnh7; Egr1; Hbb-bs; Nr4a3; Kcnv1; Gpam; H2aw; Itm2a; Slc16a4; Hbb-bs; Hba-a1; Prom1; Tspan2. |
Functional enrichment analysis of DEGs
Biological processes and KEGG annotation were applied to explore the function of DEGs. All DEGs significantly played a role in localization, signaling, metabolic process, development process, and positive regulation of biological process (Figure 2A). Twenty-four biological process terms were filtered with P value less than 0.001, and DEGs were significantly enriched in the regulation of neuronal apoptosis (Figure 2B). Next, we screened the biological processes associated with neuron death with the key words neuron and death, and 5 biological processes (positive regulation of cell death, regulation of neuron death, negative regulation of neuron death, regulation of neuron apoptotic process and negative regulation of neuron apoptotic process) were dysregulated by sevoflurane (Figure 3A). Most of the genes enriched in disordered biological processes associated with cell death were upregulated after sevoflurane inhalation (Figure 3B–3F). A total of 10 KEGG pathways, such as peroxisome, AGE-RAGE signaling pathway in diabetic complications, inositol phosphate metabolism, vascular smooth muscle contraction, rap1 signaling pathway, and glycerophospholipid metabolism, were enriched by DEGs (Figure 2C).
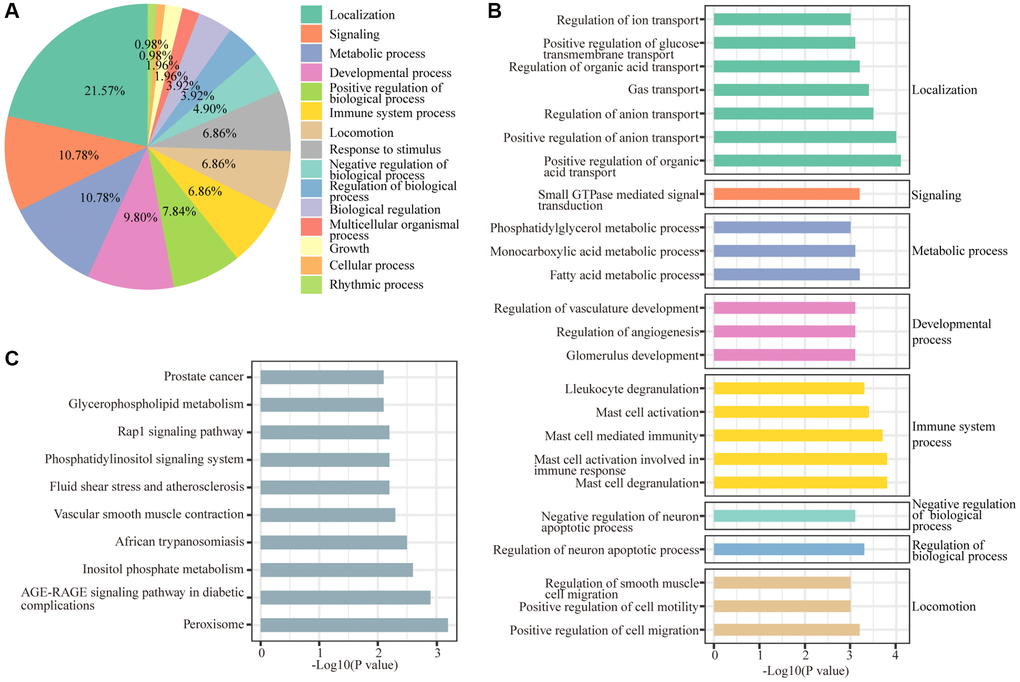
Figure 2. GO and KEGG enrichment analyses of differentially regulated genes. (A) Functions of biological processes that are significantly enriched by differentially expressed genes. (B) Top 20 significantly enriched biological processes. (C) Ten KEGG pathways were significantly enriched by differentially expressed genes.
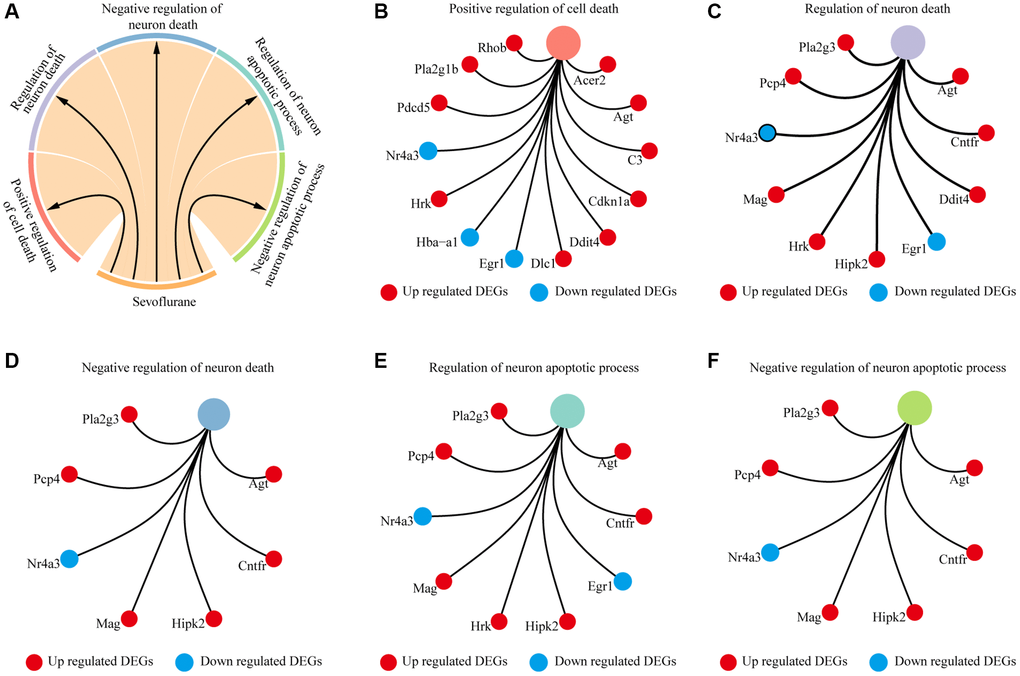
Figure 3. Cell death-related biological processes were significantly enriched by differentially expressed genes. (A) Five cell death-related biological processes were significantly enriched by differentially expressed genes. (B) Thirteen differentially expressed genes (10 genes upregulated in the sevoflurane-treated group and 3 genes downregulated in the sevoflurane-treated group) were significantly enriched in positive regulation of cell death. (C) Ten differentially expressed genes (8 genes upregulated in the sevoflurane-treated group and 2 genes downregulated in the sevoflurane-treated group) were significantly enriched in the regulation of neuronal death. (D) Seven differentially expressed genes (6 genes upregulated in the sevoflurane-treated group and 1 gene downregulated in the sevoflurane-treated group) were significantly enriched in the negative regulation of neuronal death. (E) Nine differentially expressed genes (7 genes upregulated in the sevoflurane-treated group and 2 genes downregulated in the sevoflurane-treated group) were significantly enriched in the regulation of neuronal apoptosis. (F) Seven differentially expressed genes (6 genes upregulated in the sevoflurane-treated group and 1 gene downregulated in the sevoflurane-treated group) were significantly enriched in the negative regulation of neuronal apoptosis. (Red: up regulated DEGs; blue: down regulated DEGs).
Protein-protein interaction network construction and hub gene selection
A total of 57 nodes and 97 interactions of the DEGs were identified in STRING and were visualized in Cytoscape (Figure 4). We calculated the number of genes enriched in biological process terms, and the genes that were enriched in at least 10 terms are listed in Table 2. The cytoHubba application identified 58 hub genes with 12 algorithms, including 29 genes that were identified by at least five different methods as candidate hub genes (Table 3). Six hub genes (Agt, Cdkn1a, Ddit4, Pdgfra, Rapgef3, and Rhob) were both selected with two methods (Figure 5A). The six hub genes were upregulated after sevoflurane inhalation (Figure 5B). We further validated the expression of the six hub genes in vivo. We found that 4 h of 3% sevoflurane treatment increased the mRNA levels of Agt, Cdkn1a, Ddit4, Pdgfra, Rapgef3, and Rhob in the mouse hippocampus (Figure 5C). Among the 6 hub genes, 4 genes (Agt, Cdkn1a, Ddit4, and Rhob) were also enriched in biological processes associated with neuronal death (Figure 5D).
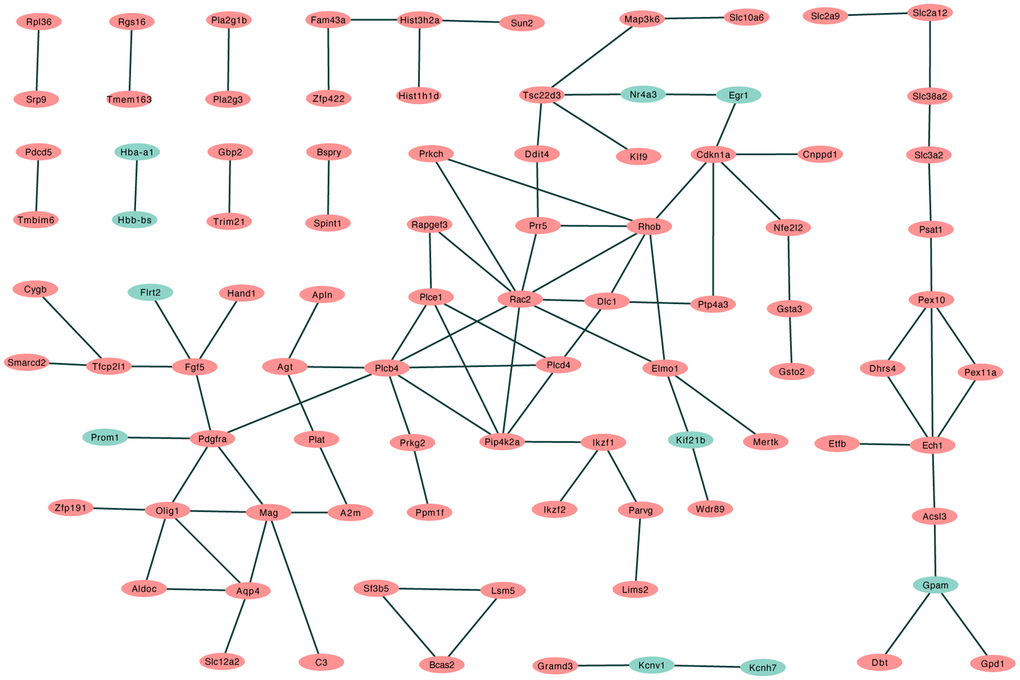
Figure 4. PPI network of differentially regulated genes in STRING. (Red: upregulated differentially expressed genes in the sevoflurane-treated group; green: downregulated differentially expressed genes in the sevoflurane-treated group).
Table 2. Number of biological process terms per gene involved.
| Genes | Terms | Genes | Terms | Genes | Terms | Genes | Terms | Genes | Terms |
| Agt | 57 | Pla2g1b | 11 | Prr5 | 6 | Mag | 4 | Ech1 | 2 |
| Nr4a3 | 40 | Gpam | 10 | Rac2 | 6 | Rab31 | 4 | Etfb | 2 |
| Pla2g3 | 34 | Sparc | 10 | Cnksr3 | 5 | Serpinb3a | 4 | Kcnh7 | 2 |
| Pdgfra | 27 | Tgfbr3 | 10 | Cntfr | 5 | Slc10a6 | 4 | Kcnv1 | 2 |
| Egr1 | 25 | Dlc1 | 9 | Fgf5 | 5 | Slc16a4 | 4 | Klf9 | 2 |
| Nfe2l2 | 22 | Plce1 | 9 | Flrt2 | 5 | Sun2 | 4 | Slc2a9 | 2 |
| Rapgef3 | 21 | Ackr3 | 8 | Mertk | 5 | Aqp4 | 3 | Trim21 | 2 |
| Hipk2 | 20 | Fxyd1 | 8 | Oprm1 | 5 | Clic1 | 3 | Aldoc | 1 |
| C3 | 17 | Pcp4 | 8 | Plat | 5 | Cygb | 3 | Cnppd1 | 1 |
| Rhob | 16 | Tnfrsf11a | 8 | Prkg2 | 5 | Ecrg4 | 3 | Elmo1 | 1 |
| Ptgds | 14 | Acsl3 | 7 | Snx9 | 5 | Hand1 | 3 | Gm266 | 1 |
| Ppm1f | 13 | Apln | 7 | Timp4 | 5 | Pip4k2a | 3 | Lims2 | 1 |
| Slc12a2 | 13 | Ddah1 | 7 | Tmbim6 | 5 | Plcb4 | 3 | Paqr8 | 1 |
| Ddit4 | 12 | Pdcd5 | 7 | A2m | 4 | Plcd4 | 3 | Peli2 | 1 |
| Adipor2 | 11 | Spint1 | 7 | Hba-a1 | 4 | Prom1 | 3 | Prlhr | 1 |
| Cdkn1a | 11 | Acer2 | 6 | Hrk | 4 | Slc38a2 | 3 | Rabif | 1 |
| Mrgprb3 | 11 | Pla2g1b | 11 | Ikzf1 | 4 | Slc3a2 | 3 | | |
Table 3. Times of hub genes selected from 12 algorithms with cytoHubba in cytoscape.
| Genes | Times | Genes | Times | Genes | Times | Genes | Times | Genes | Times |
| Ddit4 | 12 | Plcd4 | 10 | Tsc22d3 | 8 | A2m | 3 | Pla2g3 | 3 |
| Elmo1 | 12 | Plce1 | 10 | Prkch | 7 | Bcas2 | 3 | Prom1 | 3 |
| Mag | 12 | Agt | 9 | Kcnv1 | 6 | Cygb | 3 | Psat1 | 3 |
| Olig1 | 12 | Cdkn1a | 9 | Kif21b | 6 | Dbt | 3 | Sf3b5 | 3 |
| Pdgfra | 12 | Fgf5 | 9 | Ptp4a3 | 6 | Gpam | 3 | Apln | 2 |
| Ddit4 | 12 | Ikzf1 | 9 | Rapgef3 | 6 | Gsto2 | 3 | Map3k6 | 2 |
| Pip4k2a | 12 | Prkg2 | 9 | Dhrs4 | 5 | Hist3h2a | 3 | Ppm1f | 2 |
| Plcb4 | 12 | Tfcp2l1 | 9 | Egr1 | 5 | Lsm5 | 3 | Slc3a2 | 2 |
| Prr5 | 12 | Ech1 | 8 | Acsl3 | 4 | Mertk | 3 | Ikzf2 | 1 |
| Rac2 | 12 | Gsta3 | 8 | Aldoc | 4 | Parvg | 3 | Slc38a2 | 1 |
| Rhob | 12 | Pex10 | 8 | Dlc1 | 4 | Pex11a | 3 | Wdr89 | 1 |
| Aqp4 | 10 | Plat | 8 | Nfe2l2 | 4 | Pla2g1b | 3 | | |
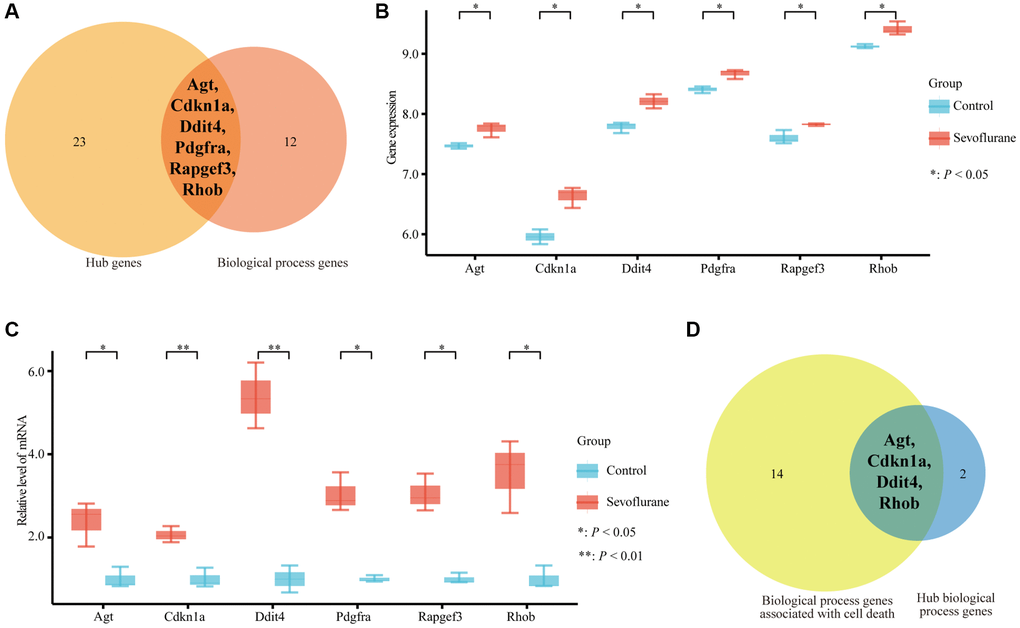
Figure 5. Six hub genes were enriched in biological process terms, and 4 hub genes were associated with cell death biological process terms. (A) Six hub genes enriched in biological process terms. (B) Expression of 6 hub genes between the control and sevoflurane-treated groups. (C) qPCR detection of Agt, Cdkn1a, Ddit4, Pdgfra, Rapgef3, and Rhob mRNA expression levels in the hippocampus of the mice that received 3% sevoflurane exposure for 4 h or the control mice. Sevoflurane indicates the mice received sevoflurane exposure. Control indicates that the mice were raised only under normal conditions. (D) Four hub genes associated with cell death biological process terms. The data shown are the means ± SDs, n = 3. *P < 0.05, **P < 0.01. (Green: Control group; Red: sevoflurane-treated group).
Sevoflurane upregulated the expression of Ddit4
DDIT4, an encoded protein that regulates development and DNA damage and participates in various pathological processes, was significantly enriched in the regulation of neuron death and positive regulation of cell death (Figure 6A). NMDAR and GABAAR are considered important targets of sevoflurane [36–38]. Therefore, we further explored whether DDIT4 is regulated by NMDAR or GABAAR. We found that activation of GABAAR by injection of the GABAAR agonist muscimol (1.25 μg) into the mouse hippocampus did not cause a significant change in Ddit4 expression (Figure 6B). However, after injection of the NMDAR antagonist MK-801 (0.25 μg) into the mouse hippocampus (injection coordinates: AP −2.1 mm, ML 1.5 mm, DV −2.1 mm) by brain stereotactic injection, the mRNA expression of Ddit4 was increased (Figure 6C).
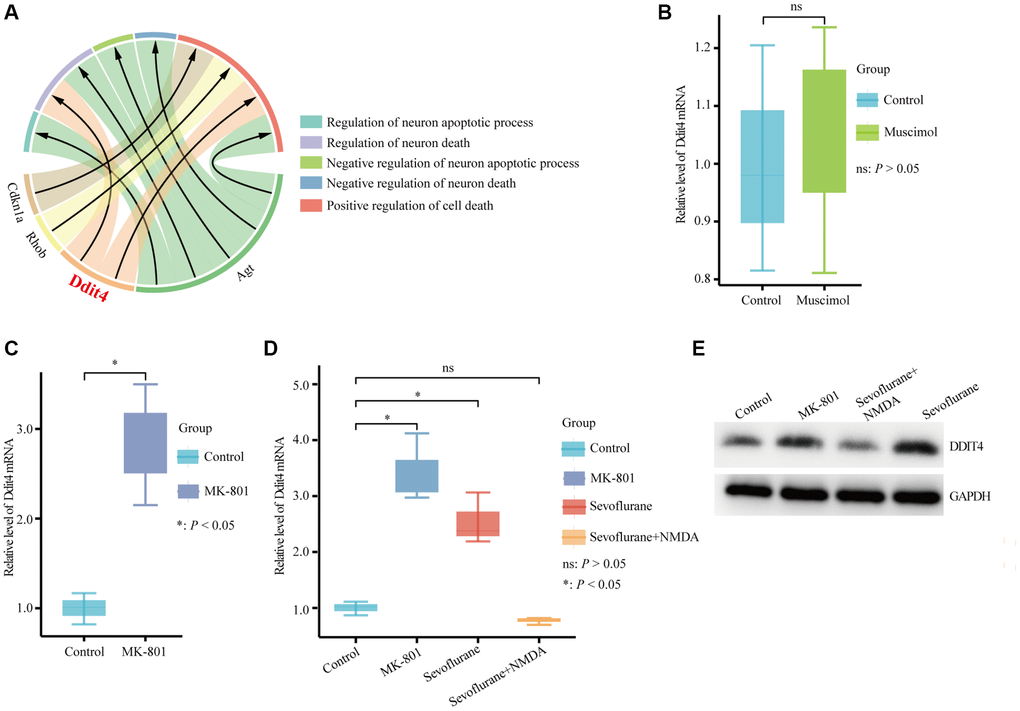
Figure 6. Hub genes associated with cell death biological process terms. (A) Four hub genes enriched in cell death biological process terms. (B) qPCR detection of the Ddit4 mRNA level in the hippocampus of mice with hippocampal stereotactic injection of muscimol or saline. (C) qPCR detection of the Ddit4 mRNA level in the hippocampus of mice with the hippocampal stereotactic injection of MK-801 or saline. (D) qPCR detection indicated the Ddit4 mRNA level in the hippocampus of mice that received sevoflurane exposure with NMDA (sevoflurane + NMDA group) or saline (sevoflurane group) injection into the hippocampus. Control indicates mice injected with saline in the hippocampus. MK-801 indicates mice injected with MK-801 in the hippocampus. (E) Representative pictures showed that DDIT4 level was elevated as shown by western blotting. The data shown are the means ± SDs, n = 3. nsP > 0.05, *P < 0.05.
While using sevoflurane for anesthesia treatment, we injected 0.5 μg NMDA into the hippocampus (AP −2.1 mm, ML 1.5 mm, DV −2.1 mm) of mice and found that the increased expression of Ddit4 caused by sevoflurane could be rescued by NMDA, indicating that the effect of sevoflurane on the expression of DDIT4 might occur through the NMDA receptor (Figure 6D). The western blot results show that DDIT4 level was elevated after sevoflurane-treated, but decrease after NMDA supplementation (Figure 6E).
Discussion
As a common perioperative neurological impairment in elderly patients, POCD strongly affects rapid recovery and long-term quality of life and places a heavy burden on patients’ families and society [39]. Neuronal apoptosis induced by sevoflurane is one of the possible factors leading to POCD [10, 40, 41]. Sevoflurane may lead to neuronal death or neuroinflammation to induce cognitive impairment [42]. In this study, we comprehensively analyzed a total of 170 DEGs, 153 upregulated genes and 17 downregulated genes, in the hippocampus of aged rats after sevoflurane anesthesia, and 4 hub genes (Agt, Cdkn1a, Ddit4, and Rhob) were critically related to the biological process of cell death. We further confirmed the upregulation of these genes, especially Ddit4, in the hippocampus of the aged mice that received 4 hours of sevoflurane anesthesia. NMDAR, the core target receptor of sevoflurane, rather than GABAAR, mediates the sevoflurane regulation of DDIT4 expression.
We screened the DEGs from the hippocampus of rats, which is closely related to cognitive function [43] and may play an important role in the pathogenesis of POCD [44, 45]. Agt encodes angiotensinogen, an angiotensin precursor protein that functions in the renin-angiotensin system (RAS). In addition to the liver, Agt is also expressed in the brain. Increasing evidence has shown that the brain RAS plays a key role in Alzheimer's disease, stroke, alcoholism, and depression [46]. Angiotensin regulates iron homeostasis in dopaminergic neurons and microglia through type 1 receptors, thus affecting neurodegenerative diseases such as Parkinson’s disease [47]. The interruption of angiotensinogen synthesis in astrocytes in the rat brain affects the function of the locus coeruleus, which may be responsible for cognitive, behavioural, and sleep disorders [48]. In this study, we found that Agt participates in both the positive and negative regulation of neuronal apoptosis. This evidence suggests that the overexpression of Agt in the hippocampus of aged rats after sevoflurane anesthesia may lead to dysfunction of the brain RAS system by affecting neuronal apoptosis.
Cdkn1a encodes cyclin-dependent kinase inhibitor 1A, which is mainly involved in cell cycle regulation. Several studies have shown that cell cycle-related molecules and pathways play a variety of important roles in influencing neuronal function. In some brain diseases, it is thought that cell cycle arrest may increase the susceptibility to cell death [49]. The failure of cell cycle regulation leads to neuronal dysfunction and cell death, which may be the underlying cause of several neurodegenerative diseases and the ultimate common pathway of other neurodegenerative diseases [50, 51]. Our study confirmed that the Cdkn1a gene is enriched in the biological process of positive regulation of cell death, and the overexpression of Cdkn1a after sevoflurane treatment may disrupt the normal cell cycle and accelerate neuronal death in the hippocampus. Similarly, the small molecule GTPase Rhob encoded by Rhob is an important regulator of cytoskeletal tissue and vesicle and membrane receptor transport. Researchers have found that RHOB is highly expressed in the hippocampus and may be essential for synaptic plasticity in the hippocampus [52]. Moreover, Rhob plays a key role in the apoptotic response, and its deletion affects the apoptotic response of tumor cells to DNA damage [53]. Therefore, both Cdkn1a and Rhob may be the possible pathological basis of sevoflurane neurotoxicity.
In this study, we found that Ddit4 is the only key gene enriched in both neuronal death and unidirectional regulation of apoptosis. Ddit4, also known as REDD1 and RTP801, encodes proteins that regulate development and DNA damage and participate in a variety of pathological processes. Suppression of DDIT4 expression decreases cell apoptosis in many kinds of cells [54–56]. Overexpression of DDIT4 promoted SUNE1 cell proliferation but inhibited apoptosis [57]. Here, we showed that sevoflurane upregulates DDIT4 expression, which suggests that neuronal apoptosis is induced by sevoflurane neurotoxicity.
The apoptosis-related neuronal death process regulated by sevoflurane leading to cognitive impairment has been recognized. Inhalation of 2% sevoflurane for 5 hours can activate the NF-κB signaling pathway and promote neuronal apoptosis and the production of inflammatory factors, thus affecting learning and memory abilities [58]. Activation of the PI3K/Akt signaling pathway reduces hippocampal neuronal apoptosis and exerts a protective effect against sevoflurane-induced brain injury in aged rats [59]. We also confirmed that 3% sevoflurane treatment increased the mRNA levels of Agt, Cdkn1a, Ddit4, Pdgfra, Rapgef3, and Rhob in the mouse hippocampus. The expression of Ddit4 in the hippocampal CA1 region was significantly altered after chronic cerebral hypoperfusion, indicating that it may play an important role in neuronal injury [60]. Inhibition of DDIT4 could reverse metformin-induced cell cycle arrest and significantly protect against the deleterious effects of the drug on cellular transformation [61]. Inhibition of DDIT4 expression also exerted a neuroprotective effect after ischemia-reperfusion injury [62]. These results suggest that DDIT4 may be a key target for intervention in cell apoptosis induced by sevoflurane.
General anesthetics play an anesthetic role mainly by inhibiting the target receptor NMDAR and activating GABAAR to regulate nerve signal transduction and can further induce a wide range of physiological effects through NMDAR and GABAAR to regulate downstream molecular signal pathways [31, 38, 63, 64]. Inhibition of NMDAR by MK801 leads to apoptosis of neurons [65, 66]. MK-801 also inhibits proliferation and increases apoptosis in hippocampal neural stem cells [67]. We found that the upregulation of DDIT4 expression in the hippocampus by sevoflurane can be inhibited through the supplementation of NMDA in the hippocampus. The injection of MK-801 into the hippocampus of mice also significantly promoted the expression of DDIT4. However, GABAAR activation did not significantly affect the regulatory effect of sevoflurane on DDIT4 expression. This finding indicates that sevoflurane regulates the expression of DDIT4 through NMDAR rather than GABAAR.
However, a limitation in our analysis was that we screened 4 hub genes while we only explored the mechanism of DDIT4 elevation after sevoflurane inhaling. Mechanisms of the change in the other three genes need to be explored in future experiments. Here we used the antagonist of NMDAR MK-801 to determine the NMDAR mediating sevoflurane regulation, the gain and loss function of NMDAR subunits by RNAi will be performed in the future. In addition, we will further perform the overexpression or knockdown of DDIT4 in the hippocampus to investigate whether Ddit4 is involved in the sevoflurane-induced neuron death.
Our study comprehensively analyzed sevoflurane-regulated DEGs to indicate that Ddit4 may be a potential target of sevoflurane-induced neuronal apoptosis and determined that the NMDAR/DDIT4 pathway may be a potential target of sevoflurane neurotoxicity, which provides new possibilities for the prevention and treatment of sevoflurane neurotoxicity.
Materials and Methods
Microarray data analysis
GSE139220 expression profiles were retrieved and obtained from the NCBI-GEO website (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139220) [68]. The whole transcriptomic data of hippocampal tissue from 3 rats that received 100% oxygen at an identical flow rate for 4 h in an identical chamber and 3 rats that received 2.5% sevoflurane in 100% oxygen for 4 hours in an anesthetizing chamber were included. The raw data were normalized with the rma function by using the oligo package on the R version 4.2.2 platform [69]. The expression data were annotated with the SwissProt database. If the target gene was annotated with two or more probes, the mean value was calculated. Then, the Limma package for the R environment was used to detect the differentially expressed genes (DEGs) in hippocampal tissue between the control group rats and the sevoflurane-treated rats [70]. DEGs were identified based on a P value less than 0.05.
DEG functional enrichment analysis
Gene enrichment analysis of DEGs was performed on the web-based portal Metascape (http://metascape.org/) [71] using the Gene Ontology biological processes and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [34]. The enrichment terms were visualized using the ggplot2 package in R.
Protein-protein interaction network construction
For all DEGs, a protein-protein interaction (PPI) network was constructed using the STRING database (https://cn.string-db.org/) [72]. Then, the network was visualized on Cytoscape software version 3.9.1, which can be freely downloaded on the website https://cytoscape.org/ and can be used to detect hub genes with the cytoHubba app [73, 74].
Hub gene selection
To explore the hub genes, we used two screening methods. One is that this gene is involved in multiple biological processes, and the other is that hub genes were screened by 12 algorithms with cytoHubba in Cytoscape, and the genes that were identified by both methods were considered to play a critical role in sevoflurane neurotoxicity. The hub genes enriched in the neuron death process were considered to be involved in neurotoxicity.
In vivo validation
We performed in vivo tests to validate the results from the microarray data. The animal experimental protocol was approved by the Standing Committee on Animals at National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (protocol number: NCC2021A586). Eighteen-month-old C57BL/6J female mice were raised in clean cages under pathogen-free conditions (22–26°C, 12/12 h light/dark cycle) and offered food and water ad libitum. We established a mouse model that received a clinical concentration of 3% sevoflurane (with 40% oxygen and 57% nitrogen) for 4 h of anesthesia (n = 3). The temperature was controlled to maintain at 30°C during anesthesia. The control group mice (n = 3) were raised under normal rearing conditions.
The NMDAR antagonist MK-801 (1 μl, 0.25 μg) (Selleck, USA), the GABAAR agonist muscimol (1 μl, 1.25 μg) (MedChemExpress, China), NMDA (1 μl, 0.5 μg) (Selleck) or 1 μl of saline was injected into the mouse hippocampus (injection coordinates: AP −2.1 mm, ML 1.5 mm, DV −2.1 mm) by brain stereotactic injection.
Quantitative real-time PCR (qPCR)
Total RNA from the hippocampus was isolated by using RNAiso Plus (TaKaRa, China). cDNA synthesis from mRNA was performed by using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa). Then, the cDNA was used for qPCR detection by using Fast qPCR Mix (TaKaRa). Primers for the qPCR analysis of mRNA are shown as follows:
Ddit4-PF: 5′-CAAGGCAAGAGCTGCCATAG-3′, Ddit4-PR: 5′-CCGGTACTTAGCGTCAGGG-3′; Pdgfra-PF: 5′-AGAGTTACACGTTTGAGCTGTC-3′, Pdgfra-PR: 5′-GTCCCTCCACGGTACTCCT-3′; Rhob-PF: 5′-GTGCCTGCTGATCGTGTTCA-3′, Rhob-PR: 5′-CCGAGAAGCACATAAGGATGAC-3′; Agt-PF: 5′-TCTCCTTTACCACAACAAGAGCA-3′, Agt-PR: 5′-CTTCTCATTCACAGGGGAGGT-3′; Cdkn1a-PF: 5′-CCTGGTGATGTCCGACCTG-3′, Cdkn1a -PR: 5′-CCATGAGCGCATCGCAATC-3′; Rapgef3-PF: 5′- TCTTACCAGCTAGTGTTCGAGC-3′, Rapgef3-PR: 5′-AATGCCGATATAGTCGCAGATG-3′.
Statistical analysis
Statistical analysis was performed on the R version 4.2.2 platform. The quantitative data are presented as the mean ± SD. The microarray data and in vivo PCR validations are displayed with boxplots. Unpaired two-tailed Student’s t-test was used to determine significant differences between the two groups. A P value less than 0.05 was considered significant.
Abbreviations
POCD: Postoperative cognitive dysfunction;
DEGs: differentially expressed genes;
Cx43: connexin 43;
GABAAR: gamma-aminobutyric acid subtype A receptor;
NMDAR: N-methyl-D-aspartate receptor;
KEGG: Kyoto Encyclopedia of Genes and Genomes;
qPCR: Real-time qRT-PCR;
RAS: renin-angiotensin system.
Author Contributions
Project design and supervision: Hui Zheng, Shuai Li. Generation of the critical concept: Shuai Li. Experience performance and data analysis: Qi Hou, Bo Zhang, Runjia Wang, Qiang Wang, Yu Hou. Writing and revision of the manuscript: Shuai Li, Cheng Ni.
Acknowledgments
We are very grateful to the providers who submitted the transcriptomic data to the public databases.
Conflicts of Interest
The authors declare no conflicts of interest related to this study.
Ethical Statement
The animal protocol was approved by the Standing Committee on Animals at National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (protocol number: NCC2021A586).
Funding
The present study was supported by the National Natural Science Foundation of China (Grant No. 82101281, Grant No. 82271233, Grant No. 81970994, Grant No. 82171195).
References
-
1.
Weiser TG, Haynes AB, Molina G, Lipsitz SR, Esquivel MM, Uribe-Leitz T, Fu R, Azad T, Chao TE, Berry WR, Gawande AA. Estimate of the global volume of surgery in 2012: an assessment supporting improved health outcomes. Lancet. 2015 (Suppl 2); 385:S11. https://doi.org/10.1016/S0140-6736(15)60806-6 [PubMed]
-
2.
Evered LA, Silbert BS. Postoperative Cognitive Dysfunction and Noncardiac Surgery. Anesth Analg. 2018; 127:496–505. https://doi.org/10.1213/ANE.0000000000003514 [PubMed]
-
3.
Urits I, Orhurhu V, Jones M, Hoyt D, Seats A, Viswanath O. Current Perspectives on Postoperative Cognitive Dysfunction in the Ageing Population. Turk J Anaesthesiol Reanim. 2019; 47:439–47. https://doi.org/10.5152/TJAR.2019.75299 [PubMed]
-
4.
Evered LA, Silbert BS, Scott DA, Maruff P, Ames D. Prevalence of Dementia 7.5 Years after Coronary Artery Bypass Graft Surgery. Anesthesiology. 2016; 125:62–71. https://doi.org/10.1097/ALN.0000000000001143 [PubMed]
-
5.
Newman MF, Kirchner JL, Phillips-Bute B, Gaver V, Grocott H, Jones RH, Mark DB, Reves JG, Blumenthal JA, and Neurological Outcome Research Group and the Cardiothoracic Anesthesiology Research Endeavors Investigators. Longitudinal assessment of neurocognitive function after coronary-artery bypass surgery. N Engl J Med. 2001; 344:395–402. https://doi.org/10.1056/NEJM200102083440601 [PubMed]
-
6.
Luo A, Yan J, Tang X, Zhao Y, Zhou B, Li S. Postoperative cognitive dysfunction in the aged: the collision of neuroinflammaging with perioperative neuroinflammation. Inflammopharmacology. 2019; 27:27–37. https://doi.org/10.1007/s10787-018-00559-0 [PubMed]
-
7.
Netto MB, de Oliveira Junior AN, Goldim M, Mathias K, Fileti ME, da Rosa N, Laurentino AO, de Farias BX, Costa AB, Rezin GT, Fortunato JJ, Giustina AD, Barichello T, et al. Oxidative stress and mitochondrial dysfunction contributes to postoperative cognitive dysfunction in elderly rats. Brain Behav Immun. 2018; 73:661–9. https://doi.org/10.1016/j.bbi.2018.07.016 [PubMed]
-
8.
Fan D, Li J, Zheng B, Hua L, Zuo Z. Enriched Environment Attenuates Surgery-Induced Impairment of Learning, Memory, and Neurogenesis Possibly by Preserving BDNF Expression. Mol Neurobiol. 2016; 53:344–54. https://doi.org/10.1007/s12035-014-9013-1 [PubMed]
-
9.
Tang C, Zheng X, Zhong Y, Chen D, Zhu Y, Wang S, Xiong L, Zhu Z. The role of TREM1 in regulating microglial polarization in sevoflurane-induced perioperative neurocognitive disorders. J Neuroimmunol. 2023; 379:578070. https://doi.org/10.1016/j.jneuroim.2023.578070 [PubMed]
-
10.
Zhong Y, Zhao P, Zhang C, Wu Z, Fang X, Zhu Z. NUDT21 relieves sevoflurane-induced neurological damage in rats by down-regulating LIMK2. Open Life Sci. 2023; 18:20220486. https://doi.org/10.1515/biol-2022-0486 [PubMed]
-
11.
Lu J, Liu Z, Zhao Y, Liu X, He W, Zhang L. FGF19 improves sevoflurane-induced cognitive dysfunction in rats through the PGC-1α/BDNF/FNDC5 pathway. Tissue Cell. 2023; 81:102012. https://doi.org/10.1016/j.tice.2022.102012 [PubMed]
-
12.
Xu Y, Gao G, Sun X, Liu Q, Li C. ATPase Inhibitory Factor 1 Is Critical for Regulating Sevoflurane-Induced Microglial Inflammatory Responses and Caspase-3 Activation. Front Cell Neurosci. 2021; 15:770666. https://doi.org/10.3389/fncel.2021.770666 [PubMed]
-
13.
Zhang L, Xue Z, Yan J, Wang J, Liu Q, Jiang H. LncRNA Riken-201 and Riken-203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif. 2019; 52:e12573. https://doi.org/10.1111/cpr.12573 [PubMed]
-
14.
Vutskits L, Xie Z. Lasting impact of general anaesthesia on the brain: mechanisms and relevance. Nat Rev Neurosci. 2016; 17:705–17. https://doi.org/10.1038/nrn.2016.128 [PubMed]
-
15.
Lemkuil BP, Head BP, Pearn ML, Patel HH, Drummond JC, Patel PM. Isoflurane neurotoxicity is mediated by p75NTR-RhoA activation and actin depolymerization. Anesthesiology. 2011; 114:49–57. https://doi.org/10.1097/ALN.0b013e318201dcb3 [PubMed]
-
16.
Jevtovic-Todorovic V, Olney JW. PRO: Anesthesia-induced developmental neuroapoptosis: status of the evidence. Anesth Analg. 2008; 106:1659–63. https://doi.org/10.1213/ane.0b013e3181731ff2 [PubMed]
-
17.
Joseph JD, Peng Y, Mak DO, Cheung KH, Vais H, Foskett JK, Wei H. General anesthetic isoflurane modulates inositol 1,4,5-trisphosphate receptor calcium channel opening. Anesthesiology. 2014; 121:528–37. https://doi.org/10.1097/ALN.0000000000000316 [PubMed]
-
18.
Hausburg MA, Banton KL, Roman PE, Salgado F, Baek P, Waxman MJ, Tanner A 2nd, Yoder J, Bar-Or D. Effects of propofol on ischemia-reperfusion and traumatic brain injury. J Crit Care. 2020; 56:281–7. https://doi.org/10.1016/j.jcrc.2019.12.021 [PubMed]
-
19.
Li G, Yu B. Elevation of protective autophagy as a potential way for preventing developmental neurotoxicity of general anesthetics. Med Hypotheses. 2014; 82:177–80. https://doi.org/10.1016/j.mehy.2013.11.032 [PubMed]
-
20.
Li M, Guo J, Wang H, Li Y. Involvement of Mitochondrial Dynamics and Mitophagy in Sevoflurane-Induced Cell Toxicity. Oxid Med Cell Longev. 2021; 2021:6685468. https://doi.org/10.1155/2021/6685468 [PubMed]
-
21.
Xu M, Feng J, Tang M, Guo Q, Zhan J, Zhu F, Lei H, Kang Q. Blocking retrograde axonal transport of autophagosomes contributes to sevoflurane-induced neuron apoptosis in APP/PS1 mice. Acta Neurol Belg. 2021; 121:1207–15. https://doi.org/10.1007/s13760-020-01359-6 [PubMed]
-
22.
Wang Q, Li Y, Tan H, Wang Y. Sevoflurane-Induced Apoptosis in the Mouse Cerebral Cortex Follows Similar Characteristics of Physiological Apoptosis. Front Mol Neurosci. 2022; 15:873658. https://doi.org/10.3389/fnmol.2022.873658 [PubMed]
-
23.
Zhang L, Mao H, Yan J, Cheng Y, Xue Z, Qiu Z, Jiang H. Sevoflurane enhances brain glycolysis and lactate production in aged marmosets. Br J Anaesth. 2022; 129:e63–6. https://doi.org/10.1016/j.bja.2022.05.035 [PubMed]
-
24.
Li Q, Qin X, Kou X, Li J, Li Z, Chen C. Anagliptin promotes apoptosis in mouse colon carcinoma cells via MCT-4/lactate-mediated intracellular acidosis. Exp Ther Med. 2022; 23:282. https://doi.org/10.3892/etm.2022.11211 [PubMed]
-
25.
Liu Q, Ruan H, Sheng Z, Sun X, Li S, Cui W, Li C. Nanoantidote for repression of acidosis pH promoting COVID-19 infection. View (Beijing). 2022; 3:20220004. https://doi.org/10.1002/VIW.20220004 [PubMed]
-
26.
Li J, Chen L, Qin Q, Wang D, Zhao J, Gao H, Yuan X, Zhang J, Zou Y, Mao Z, Xiong Y, Min Z, Yan M, et al. Upregulated hexokinase 2 expression induces the apoptosis of dopaminergic neurons by promoting lactate production in Parkinson's disease. Neurobiol Dis. 2022; 163:105605. https://doi.org/10.1016/j.nbd.2021.105605 [PubMed]
-
27.
Wei K, Liu Y, Yang X, Liu J, Li Y, Deng M, Wang Y. Bumetanide attenuates sevoflurane-induced neuroapoptosis in the developing dentate gyrus and impaired behavior in the contextual fear discrimination learning test. Brain Behav. 2022; 12:e2768. https://doi.org/10.1002/brb3.2768 [PubMed]
-
28.
Dong Y, Zhang G, Zhang B, Moir RD, Xia W, Marcantonio ER, Culley DJ, Crosby G, Tanzi RE, Xie Z. The common inhalational anesthetic sevoflurane induces apoptosis and increases beta-amyloid protein levels. Arch Neurol. 2009; 66:620–31. https://doi.org/10.1001/archneurol.2009.48 [PubMed]
-
29.
Liu F, Rainosek SW, Frisch-Daiello JL, Patterson TA, Paule MG, Slikker W Jr, Wang C, Han X. Potential Adverse Effects of Prolonged Sevoflurane Exposure on Developing Monkey Brain: From Abnormal Lipid Metabolism to Neuronal Damage. Toxicol Sci. 2015; 147:562–72. https://doi.org/10.1093/toxsci/kfv150 [PubMed]
-
30.
Lu Y, Wu X, Dong Y, Xu Z, Zhang Y, Xie Z. Anesthetic sevoflurane causes neurotoxicity differently in neonatal naïve and Alzheimer disease transgenic mice. Anesthesiology. 2010; 112:1404–16. https://doi.org/10.1097/ALN.0b013e3181d94de1 [PubMed]
-
31.
Wang WY, Jia LJ, Luo Y, Zhang HH, Cai F, Mao H, Xu WC, Fang JB, Peng ZY, Ma ZW, Chen YH, Zhang J, Wei Z, et al. Location- and Subunit-Specific NMDA Receptors Determine the Developmental Sevoflurane Neurotoxicity Through ERK1/2 Signaling. Mol Neurobiol. 2016; 53:216–30. https://doi.org/10.1007/s12035-014-9005-1 [PubMed]
-
32.
Bi C, Cai Q, Shan Y, Yang F, Sun S, Wu X, Liu H. Sevoflurane induces neurotoxicity in the developing rat hippocampus by upregulating connexin 43 via the JNK/c-Jun/AP-1 pathway. Biomed Pharmacother. 2018; 108:1469–76. https://doi.org/10.1016/j.biopha.2018.09.111 [PubMed]
-
33.
Jing Q, Zhang H, Sun X, Xu Y, Cao S, Fang Y, Zhao X, Li C. A Comprehensive Analysis Identified Hub Genes and Associated Drugs in Alzheimer's Disease. Biomed Res Int. 2021; 2021:8893553. https://doi.org/10.1155/2021/8893553 [PubMed]
-
34.
Zhang H, Cao S, Xu Y, Sun X, Fei M, Jing Q, Xu X, Tang J, Niu B, Li C. Landscape of immune infiltration in entorhinal cortex of patients with Alzheimer's disease. Front Pharmacol. 2022; 13:941656. https://doi.org/10.3389/fphar.2022.941656 [PubMed]
-
35.
Chen X, Shi L, Zhang L, Cheng Y, Xue Z, Yan J, Jiang H. Epitranscriptomic Analysis of N6-methyladenosine in Infant Rhesus Macaques after Multiple Sevoflurane Anesthesia. Neuroscience. 2022; 482:64–76. https://doi.org/10.1016/j.neuroscience.2021.11.030 [PubMed]
-
36.
Stucke AG, Zuperku EJ, Tonkovic-Capin V, Krolo M, Hopp FA, Kampine JP, Stuth EA. Sevoflurane depresses glutamatergic neurotransmission to brainstem inspiratory premotor neurons but not postsynaptic receptor function in a decerebrate dog model. Anesthesiology. 2005; 103:50–6. https://doi.org/10.1097/00000542-200507000-00011 [PubMed]
-
37.
Petrenko AB, Yamakura T, Sakimura K, Baba H. Defining the role of NMDA receptors in anesthesia: are we there yet? Eur J Pharmacol. 2014; 723:29–37. https://doi.org/10.1016/j.ejphar.2013.11.039 [PubMed]
-
38.
Lin D, Liu J, Florveus A, Ganesan V, Cottrell JE, Kass IS. Exposure to Sevoflurane, But Not Ketamine, During Early-life Brain Development has Long-Lasting Effects on GABA(A) Receptor Mediated Inhibitory Neurotransmission. Neuroscience. 2021; 472:116–27. https://doi.org/10.1016/j.neuroscience.2021.08.001 [PubMed]
-
39.
Rundshagen I. Postoperative cognitive dysfunction. Dtsch Arztebl Int. 2014; 111:119–25. https://doi.org/10.3238/arztebl.2014.0119 [PubMed]
-
40.
Liang J, Han S, Ye C, Zhu H, Wu J, Nie Y, Chai G, Zhao P, Zhang D. Minocycline Attenuates Sevoflurane-Induced Postoperative Cognitive Dysfunction in Aged Mice by Suppressing Hippocampal Apoptosis and the Notch Signaling Pathway-Mediated Neuroinflammation. Brain Sci. 2023; 13:512. https://doi.org/10.3390/brainsci13030512 [PubMed]
-
41.
Zhao J, Zhang W, Wang S, Li Z, Huang Y, Li L. Sevoflurane-induced POCD-associated exosomes delivered miR-584-5p regulates the growth of human microglia HMC3 cells through targeting BDNF. Aging (Albany NY). 2022; 14:9890–907. https://doi.org/10.18632/aging.204398 [PubMed]
-
42.
Zhou Q, Zheng Z, Wang X, Li W, Wang L, Yin C, Zhang Q, Wang Q. taVNS Alleviates Sevoflurane-Induced Cognitive Dysfunction in Aged Rats Via Activating Basal Forebrain Cholinergic Neurons. Neurochem Res. 2023; 48:1848–63. https://doi.org/10.1007/s11064-023-03871-6 [PubMed]
-
43.
Anacker C, Hen R. Adult hippocampal neurogenesis and cognitive flexibility - linking memory and mood. Nat Rev Neurosci. 2017; 18:335–46. https://doi.org/10.1038/nrn.2017.45 [PubMed]
-
44.
Liu Q, Sun YM, Huang H, Chen C, Wan J, Ma LH, Sun YY, Miao HH, Wu YQ. Sirtuin 3 protects against anesthesia/surgery-induced cognitive decline in aged mice by suppressing hippocampal neuroinflammation. J Neuroinflammation. 2021; 18:41. https://doi.org/10.1186/s12974-021-02089-z [PubMed]
-
45.
Chen L, Dong R, Lu Y, Zhou Y, Li K, Zhang Z, Peng M. MicroRNA-146a protects against cognitive decline induced by surgical trauma by suppressing hippocampal neuroinflammation in mice. Brain Behav Immun. 2019; 78:188–201. https://doi.org/10.1016/j.bbi.2019.01.020 [PubMed]
-
46.
Phillips MI, de Oliveira EM. Brain renin angiotensin in disease. J Mol Med (Berl). 2008; 86:715–22. https://doi.org/10.1007/s00109-008-0331-5 [PubMed]
-
47.
Garrido-Gil P, Rodriguez-Pallares J, Dominguez-Meijide A, Guerra MJ, Labandeira-Garcia JL. Brain angiotensin regulates iron homeostasis in dopaminergic neurons and microglial cells. Exp Neurol. 2013; 250:384–96. https://doi.org/10.1016/j.expneurol.2013.10.013 [PubMed]
-
48.
Ogier M, Bricca G, Bader M, Bezin L. Locus Coeruleus Dysfunction in Transgenic Rats with Low Brain Angiotensinogen. CNS Neurosci Ther. 2016; 22:230–7. https://doi.org/10.1111/cns.12488 [PubMed]
-
49.
Yoshizawa T, Yamagishi Y, Koseki N, Goto J, Yoshida H, Shibasaki F, Shoji S, Kanazawa I. Cell cycle arrest enhances the in vitro cellular toxicity of the truncated Machado-Joseph disease gene product with an expanded polyglutamine stretch. Hum Mol Genet. 2000; 9:69–78. https://doi.org/10.1093/hmg/9.1.69 [PubMed]
-
50.
Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007; 8:368–78. https://doi.org/10.1038/nrn2124 [PubMed]
-
51.
Nagy Z. Cell cycle regulatory failure in neurones: causes and consequences. Neurobiol Aging. 2000; 21:761–9. https://doi.org/10.1016/s0197-4580(00)00223-2 [PubMed]
-
52.
O'Kane EM, Stone TW, Morris BJ. Activation of Rho GTPases by synaptic transmission in the hippocampus. J Neurochem. 2003; 87:1309–12. https://doi.org/10.1046/j.1471-4159.2003.02102.x [PubMed]
-
53.
Liu Ax, Cerniglia GJ, Bernhard EJ, Prendergast GC. RhoB is required to mediate apoptosis in neoplastically transformed cells after DNA damage. Proc Natl Acad Sci U S A. 2001; 98:6192–7. https://doi.org/10.1073/pnas.111137198 [PubMed]
-
54.
Li L, Xi L, Wu J, Zhao Z, Chen Y, Liu W, Pan Z, Liu M, Yang D, Chen Z, Fang Y. The regulatory roles of DDIT4 in TDCIPP-induced autophagy and apoptosis in PC12 cells. J Environ Sci (China). 2023; 125:823–30. https://doi.org/10.1016/j.jes.2022.02.046 [PubMed]
-
55.
Yang J, Zhang Y, Tong J, Lv H, Zhang C, Chen ZJ. Dysfunction of DNA damage-inducible transcript 4 in the decidua is relevant to the pathogenesis of preeclampsia. Biol Reprod. 2018; 98:821–33. https://doi.org/10.1093/biolre/ioy038 [PubMed]
-
56.
Wang B, Peng L, Ouyang H, Wang L, He D, Zhong J, Xiao Y, Deng Y, Li M, Li S, Yuan J. Induction of DDIT4 Impairs Autophagy Through Oxidative Stress in Dry Eye. Invest Ophthalmol Vis Sci. 2019; 60:2836–47. https://doi.org/10.1167/iovs.19-27072 [PubMed]
-
57.
Zhao J, Li B, Ren Y, Liang T, Wang J, Zhai S, Zhang X, Zhou P, Zhang X, Pan Y, Gao F, Zhang S, Li L, et al. Histone demethylase KDM4A plays an oncogenic role in nasopharyngeal carcinoma by promoting cell migration and invasion. Exp Mol Med. 2021; 53:1207–17. https://doi.org/10.1038/s12276-021-00657-0 [PubMed]
-
58.
Tian Y, Guo S, Wu X, Ma L, Zhao X. Minocycline alleviates sevoflurane-induced cognitive impairment in aged rats. Cell Mol Neurobiol. 2015; 35:585–94. https://doi.org/10.1007/s10571-014-0154-6 [PubMed]
-
59.
Yang LH, Xu YC, Zhang W. Neuroprotective effect of CTRP3 overexpression against sevoflurane anesthesia-induced cognitive dysfunction in aged rats through activating AMPK/SIRT1 and PI3K/AKT signaling pathways. Eur Rev Med Pharmacol Sci. 2020; 24:5091–100. https://doi.org/10.26355/eurrev_202005_21202 [PubMed]
-
60.
Park JA, Lee CH. Time-Course Change of Redd1 Expressions in the Hippocampal CA1 Region Following Chronic Cerebral Hypoperfusion. Cell Mol Neurobiol. 2017; 37:563–9. https://doi.org/10.1007/s10571-016-0385-9 [PubMed]
-
61.
Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P, Tanti JF, Giorgetti-Peraldi S, Bost F. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res. 2011; 71:4366–72. https://doi.org/10.1158/0008-5472.CAN-10-1769 [PubMed]
-
62.
Wu XM, Qian ZM, Zhu L, Du F, Yung WH, Gong Q, Ke Y. Neuroprotective effect of ligustilide against ischaemia-reperfusion injury via up-regulation of erythropoietin and down-regulation of RTP801. Br J Pharmacol. 2011; 164:332–43. https://doi.org/10.1111/j.1476-5381.2011.01337.x [PubMed]
-
63.
Liu Q, Sheng Z, Cheng C, Zheng H, Lanuti M, Liu R, Wang P, Shen Y, Xie Z. Anesthetic Propofol Promotes Tumor Metastasis in Lungs via GABA(A) R-Dependent TRIM21 Modulation of Src Expression. Adv Sci (Weinh). 2021; 8:e2102079. https://doi.org/10.1002/advs.202102079 [PubMed]
-
64.
Sebel LE, Richardson JE, Singh SP, Bell SV, Jenkins A. Additive effects of sevoflurane and propofol on gamma-aminobutyric acid receptor function. Anesthesiology. 2006; 104:1176–83. https://doi.org/10.1097/00000542-200606000-00012 [PubMed]
-
65.
Zhou L, Welsh AM, Chen D, Koliatsos VE. NMDA inhibitors cause apoptosis of pyramidal neurons in mature piriform cortex: evidence for a nitric oxide-mediated effect involving inhibitory interneurons. Neuropharmacology. 2007; 52:1528–37. https://doi.org/10.1016/j.neuropharm.2007.02.013 [PubMed]
-
66.
Bender C, Rassetto M, de Olmos JS, de Olmos S, Lorenzo A. Involvement of AMPA/kainate-excitotoxicity in MK801-induced neuronal death in the retrosplenial cortex. Neuroscience. 2010; 169:720–32. https://doi.org/10.1016/j.neuroscience.2010.05.007 [PubMed]
-
67.
Ding J, Shao Y, Zhou HH, Ma QR, Zhang YW, Ding YX, He YQ, Liu J. Effect of NMDA on proliferation and apoptosis in hippocampal neural stem cells treated with MK-801. Exp Ther Med. 2018; 16:1137–42. https://doi.org/10.3892/etm.2018.6346 [PubMed]
-
68.
Song SY, Meng XW, Xia Z, Liu H, Zhang J, Chen QC, Liu HY, Ji FH, Peng K. Cognitive impairment and transcriptomic profile in hippocampus of young mice after multiple neonatal exposures to sevoflurane. Aging (Albany NY). 2019; 11:8386–417. https://doi.org/10.18632/aging.102326 [PubMed]
-
69.
Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010; 26:2363–7. https://doi.org/10.1093/bioinformatics/btq431 [PubMed]
-
70.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. https://doi.org/10.1093/nar/gkv007 [PubMed]
-
71.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019; 10:1523. https://doi.org/10.1038/s41467-019-09234-6 [PubMed]
-
72.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–8. https://doi.org/10.1093/nar/gkw937 [PubMed]
-
73.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–504. https://doi.org/10.1101/gr.1239303 [PubMed]
-
74.
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014 (Suppl 4); 8:S11. https://doi.org/10.1186/1752-0509-8-S4-S11 [PubMed]