New perspectives on the potential of tetrandrine in the treatment of non-small cell lung cancer: bioinformatics, Mendelian randomization study and experimental investigation
Abstract
Background: Although there are numerous treatment methods for NSCLC, long-term survival remains a challenge for patients. The objective of this study is to investigate the role and causal relationship between the target of tetrandrine and non-small cell lung cancer (NSCLC) through transcriptome and single-cell sequencing data, summary-data-based Mendelian Randomization (SMR) and basic experiments. The aim is to provide a new perspective for the treatment of NSCLC.
Methods: We obtained the drug target gene of tetrandrine through the drug database, and then used the GSE19188 data set to obtain the NSCLC pathogenic gene, established a drug-disease gene interaction network, screened out the hub drug-disease gene, and performed bioinformatics and tumor cell immune infiltration analysis. Single-cell sequencing data (GSE148071) to determine gene location, SMR to clarify causality and drug experiment verification.
Results: 10 drug-disease genes were obtained from 213 drug targets and 529 disease genes. DO/GO/KEGG analysis showed that the above genes were all related to the progression and invasion of NSCLC. Four drug-disease genes were identified from a drug-disease PPI network. These four genes were highly expressed in tumors and positively correlated with plasma cells, T cells, and macrophages. Subsequent single-cell sequencing data confirmed that these four genes were distributed in epithelial cells, and SMR analysis revealed the causal relationship between CCNA2 and CCNB1 and the development of NSCLC. The final molecular docking and drug experiments showed that CCNA2 and CCNB1 are key targets for tetrandrine in the treatment of NSCLC.
Introduction
According to the 2018-2020 global cancer statistics [1, 2], Among all malignant tumors, lung cancer has one of the highest incidence rates (over 10%) and the highest mortality rate (over 18.4%). Non-small cell lung cancer (NSCLC) is the predominant pathological type of lung cancer, which includes adenocarcinoma, squamous cell carcinoma, and large cell carcinoma [3, 4]. Surgery, radiation therapy, chemotherapy, targeted therapy, and immunotherapy are among the primary treatment options for NSCLC. Early NSCLC can usually be treated with surgery and stereotactic radiotherapy, but clinically most patients are diagnosed at an advanced stage and have lost the chance of radical cure. In recent years, targeted therapy has made important progress in non-squamous non-small cell lung cancer, especially the efficacy of targeted drugs targeting mutated genes such as EGFR, ALK, ROS1, and BRAF has been fully confirmed [5]. At the same time, with the widespread application of immune checkpoint inhibitors in the treatment of NSCLC, the overall survival of a considerable number of patients has been prolonged [6]. However, for most NSCLC patients, drug resistance is an unavoidable problem with the above therapies, so researchers need to find other treatment strategies.
Traditional Chinese medicine has a long history in the treatment of non-small cell lung cancer, and has accumulated rich clinical experience. CATLA study showed that the Chinese herbal medicine Yiqi Yangyin Jiedu Decoction (YYJD) can delay the occurrence of acquired resistance to the first-generation EGFR-TKI, which provides a new idea for the treatment of non-small cell lung cancer [7]. Studies have found that theabrownin (TB) is a pigment active substance in green tea, which has the ability to phosphorylate and activate MAPK/JNK pathway-related proteins, and at the same time inhibit epithelial-mesenchymal transition-related genes and pro-apoptotic molecules, p53 signaling pathway and MAPK/JNK signaling pathway, thereby effectively inhibiting the growth of NSCLC cells in xenograft models [8]. Cinobufacin (CNB) is a cardiotonic steroid or bufonilide extracted from Bufonis, which has detoxification, swelling, and analgesic effects. Studies have found that cinobufacin induces FOXO1 by inhibiting non-small cell lung cancer A549 cells G9a Regulates apoptosis, proliferation, migration and invasion [9].
Stephania tetrandra S. Moore is a traditional Chinese medicinal material. Its root is used as medicine to reduce edema and relieve pain. It can be used for dysuria and hypertension [10]. Tetrandrine is indicated for rheumatic pain, arthralgia, neuralgia, silicosis, and it can also be used in small doses in lung cancer radiotherapy [11]. Through the action of tetrandrine, the acidic environment of lysosomes is neutralized, thereby affecting the degradation process of the autophagy pathway, and at the same time inducing the apoptosis of prostate cancer, liver cancer, kidney cancer and bladder cancer cells [12]. Studies have found that tetrandrine can upregulate the mRNA and protein levels of BMP9 in colon cancer cells, and finally inhibit the proliferation of colon cancer cells through BMP9/PTEN/PI3K/AKT signaling [13]. It is reported that tetrandrine can enhance the expression of PARP, Bax, ICAM-1 and VEGF, and effectively inhibit the growth and induce apoptosis of A549 lung cancer cells by activating the VEGF/HIF-1α pathway [14]. It can be seen from the above that tetrandrine has a wide range of anticancer effects.
Variations in an individual’s genome can affect drug metabolism, absorption, distribution, and excretion, thereby affecting drug efficacy and adverse effects [15]. Therefore, knowing an individual’s drug-disease genetic information can help doctors better choose drugs and drug dosages to improve treatment efficacy and reduce the risk of adverse reactions. In addition, drug-disease genetic information can also be used to develop more personalized treatment strategies and drugs. To assess the potential efficacy of tetrandrine in NSCLC, we constructed an interaction network between tetrandrine drug targets and NSCLC oncogenes. Through screening, we identified four hub genes and investigated their relevance to immune cells. Leveraging single-cell sequencing data, we elucidated the drug’s cell-type specificity and employed SMR analysis to establish the causal relationship between these genes and NSCLC. Our findings confirmed the ability of tetrandrine to inhibit proliferation of NSCLC tumor cells and identified important targets that can inform current clinical treatments for NSCLC.
Materials and Methods
Drug target acquisition and functional analysis
From Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/) [16], SwissTargetPrediction (http://www.swisstargetprediction.ch/) [17], BindingDB (http://bindingdb.org/bind/) [18], TargetNet (http://targetnet.scbdd.com/home/index/) [19] four databases downloaded the target information of tetrandrine. In order to standardize and harmonize information obtained from various databases, we have retained the drug gene names from these database results while eliminating extraneous expression data. We then used a Venn diagram to display the combined results of drug genes in the above database. Disease Ontology (DO)/Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) analyzes were performed using these pharmacogenes, respectively.
NSCLC data obtain and analysis
NSCLC patient information and transcriptome data come from the GSE19188 dataset (https://www.ncbi.nlm.nih.gov/geo/) [20]. The dataset encompasses genetic data from a cohort of 91 patients afflicted with NSCLC, comprising 91 tumor specimens and associated information from 65 neighboring normal lung tissue samples. R was used to normalized the transcriptome data in this data set, and then the “limma” R package was used to screen the differential genes in the disease according to the standard of│logFC│>1, P<0.05. The “WGCNA” R package was used to cluster genes with similar expression patterns in GSE19188 to form different modules, and the relationship between genes and phenotypes or traits in the modules was analyzed. Finally, the intersection of the differential gene and the MEturquoise module genes analyzed by WGCNA was used as the disease gene. GO and KEGG enrichment analysis of disease genes to elucidate their functions in NSCLC.
Drug-disease joint analysis
Put all the drug and disease genes through the STRING database (https://string-db.org/) [21] to create a protein-protein interaction (PPI) network with a confidence interval of 0.4, and delete independent genes that do not interact with other genes. Take the intersection of drug gene and disease gene as drug-disease gene. Then, use Cytoscape version 3.8.0 to process the PPI network, and use MCODE to set the Degree cutoff value to 2, and the rest to the default settings to get a subnetwork consisting of 124 genes. In order to further screen hub genes, a core sub-network composed of 41 genes was screened again through the STRING database and MCODE in Cytoscape according to the above criteria. DO/GO/KEGG analyzes were performed using these hub genes, respectively. At the same time, R was used to describe the expression of 41 genes in tumor and normal tissues in NSCLC. The “ pROC “ R package established receiver operating characteristic (ROC) curves to verify the performance of hub genes.
Immune infiltration analysis
We used the CIBERSORT algorithm to analyze the level of immune cell infiltration in normal and tumor tissues in the GSE19188 dataset. In addition, we also evaluated the correlation between each hub gene and immune cells using the “IOBR” R package to determine whether these genes could be used as new biomarkers in immunotherapy. These measures help to gain insight into the immune signature of tumors and provide the basis for personalized treatment of immunotherapy.
Drug-disease gene and single cell sequencing analysis
The single-cell sequencing data set (GSE148071) was obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) [22], the dataset comprises 42 tumor samples from non-small cell lung cancer patients obtained through methods such as percutaneous biopsy, bronchoscopy, or superficial lymph node biopsy, along with their associated single-cell sequencing data. Then the single-cell data were processed using R packages such as “Seurat”, “harmony”, “UCell”, “irGSEA”, “GSVA”, and “GSEABase”. Percentages of mitochondria and rRNA are calculated by the PercentageFeatureSet function and ensure expression of more than 200 genes and <10,000 genes per cell with mitochondrial content <20%. All gene expression levels were processed by the “LogNormalize” function, and then the top 2000 highly variable genes were screened out using “FindVariableFeatures” function. After normalizing all genes, Principal Component Analysis (PCA) and Uniform Manifold Approximation and Projection (UMAP) were used for data dimensionality reduction. Cell annotation via HumanPrimaryCellAtlasData. Finally, according to the standard of min.pct = 0.25, logfc.threshold = 0.25, the differentially expressed genes are retained and combined with the drug-disease gene list to display the distribution map to see which cell subsets the drug-disease gene acts on.
Summary-data-based Mendelian randomization
This study is based on publicly available summary-level data from genome-wide association studies (GWAS) and expression quantitative trait loci (eQTL) studies. These studies have been approved by relevant institutional review boards and informed consent has been obtained from the participants. Four hub genes (CCNB1, CCNA2, BIRC5, and AURKB) were selected as exposures, and their available eQTLs were used as genetic instruments for each gene. The eQTL summary-level data were sourced from eQTLGen (https://www.eqtlgen.org/) [23]. We identified common eQTLs (minor allele frequency [MAF] >1%, P < 5.0 × 10-8) significantly associated with the expression of the four genes in blood samples. The summary-level data for non-small cell lung cancer (NSCLC) GWAS were obtained from FINNGEN (https://www.finngen.fi/en) [24] R8, with a total of 263,448 samples, including 3,865 NSCLC cases and 259,583 controls. Subsequently, SMR software version 1.31 (https://yanglab.westlake.edu.cn/software/smr/#Download) [25] was used for allele harmonization and analysis. We utilized the F-statistic to assess weak instrument variable effects and included SNPs with F > 10 to minimize weak instrument bias. For the SMR method, a P-value ≥0.01 from the heterogeneity in dependent instruments (HEIDI) test indicates no heterogeneity [26]. This step was performed to ensure the reliability and stability of the results.
Validation of all cell lines
Origin and Acquisition: The A549 and H1299 cell lines were acquired on April 5, 2023 from Dr. Mo’s laboratory. Testing and Authentication: Both the A549 and H1299 cell lines have undergone rigorous testing and authentication to ensure their identity and purity. The authentication process involved the use of standard molecular biology techniques. Method of Testing: The authentication of the A549 and H1299 cell lines was conducted using DNA profiling, specifically through short tandem repeat (STR) analysis. This method allows for the comparison of the cell line’s STR profile with known reference profiles to confirm its identity. Last Testing and Verification: The A549 and H1299 cell lines were last tested and verified for authentication on May 30, 2023. This recent authentication confirms the integrity and fidelity of the cell lines used in this study. We affirm that the above information accurately reflects the origin, authentication, testing method, and recent verification status of the A549 and H1299 cell lines used in our research.
Cytotoxicity, apoptosis assay and Edu incorporation assay
The NSCLC cell lines A549 and H1299 were maintained in 1640 medium (Hyclone, USA) containing 10% FBS (Gibco, USA) at 37° C in an atmosphere of 5% CO2. These cells were digested with pancreatin (Life-iLab, Shanghai, China). NSCLC cells were primarily seeded in several 96-well plates, and after tetrandrine treatment, CCK-8 reagent was added and operated according to the manufacturer’s instructions. Cell apoptosis was conducted by Flow cytometry using Annexin V– FITC Detection Kit (Solarbio, Beijing, China). EdU incorporation assay was employed to conduct the cell proliferation capacity of NSCLC cells treated with tetrandrine (Abbkine, China) according to the manufacturer’s instructions. Finally, images were obtained and preserved under the confocal microscope.
Molecular docking
First use the punchem website (https://pubchem.ncbi.nlm.nih.gov/) [27] to obtain the 2D structure of tetrandrine, then use ChemBio3D Ultra 14.0.0.117 to convert it into a 3D structure. The protein structure of CCNA2 and CCNB1 were obtained from the RCSB database [28]. AutoDockTools 1.5.6 (https://autodock.scripps.edu/) was utilized to prepare the receptor protein and ligand molecules, including hydrogen addition, partial charge allocation, and protonation state assignment. During molecular docking, the XYZ grid coordinates of CCNA2 were set to 18.553, 62.684, and 77.232, and the XYZ grid coordinates of CCNB1 were set to -60.067, 31.797, and -14.94, with a grid length of 40Å in each direction. The binding modes of the ligand molecules were generated using Autodock Vina [29], with a exhaustiveness setting of 8, maximum allowable energy range of 5 kcal/mol, and maximum output conformations of 20. The Protein-Ligand Interaction Profiler online site [30] is used to analyze non-covalent interactions between molecules and their ligands. PyMOL was used to process the resulting plots.
Results
Functional enrichment analysis of drug targets
213 drug target genes were identified from four drug target databases, including CTD, SwissTargetPrediction, Binding DB, and TargetNet, and their intersection was shown in Figure 1A. The results of DO analysis indicated that these drug genes mainly functioned in various cancers, such as non-small cell lung cancer, connective tissue cancer, and musculoskeletal system cancer (Figure 1B). KEGG analysis revealed that these genes were enriched in tumor pathways closely related to tumor pathways, including EGFR tyrosine kinase inhibitor resistance, PI3K-Akt signaling pathway, PD-L1 expression, and PD-1 checkpoint pathway (Figure 1C). Furthermore, GO analysis showed that drug genes were mainly enriched in biological processes of peptide-serine/threonine modification (BP), protein kinase complex in cellular components (CC), and protein serine/threonine kinase activity in molecular function (MF) (Figure 1D).
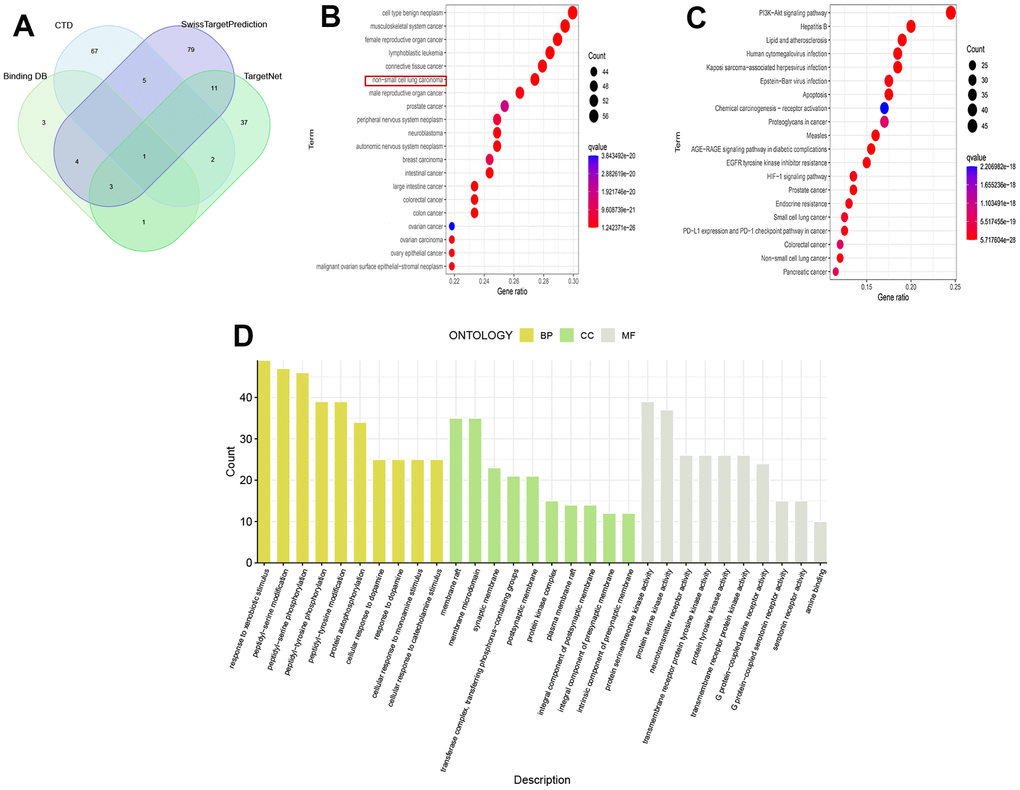
Figure 1. Analysis of drug targets. (A) Venn diagram of drug targets; (B) DO analysis; (C) KEGG analysis; (D) GO analysis.
Acquisition and analysis of disease genes
From the NSCLC data set (GSE19188), we screened out 1612 differentially expressed genes compared with normal tissues. Then, we selected a correlation coefficient of 0.86 corresponding to a soft threshold of 3 to construct a scale-free network for WGCNA analysis (Figure 2A, 2B). Through WGCNA analysis, all genes of NSCLC were constructed into 6 different gene modules, namely MEyellow, MEblue, MEturquoise, MEbrown, MEgreen and MEgrey, as shown in Figure 2C. According to the correlation analysis between the gene modules and Tumor shown in Figure 2C, These gene modules are related to the occurrence and development of NSCLC, among which MEturquoise has the most significant correlation with NSCLC and is considered as a key gene module. Based on the intersection of genes in this module and differentially expressed genes, 529 disease-causing genes were found. These pathogenic genes were mainly enriched in organelle fission (BP), chromosomal region (CC) and catalytic activity acting on DNA (MF) in functional enrichment analysis. At the same time, KEGG analysis showed that these pathogenic genes were closely related to cell cycle, DNA replication and P53 signaling pathway (Figure 2D–2F).
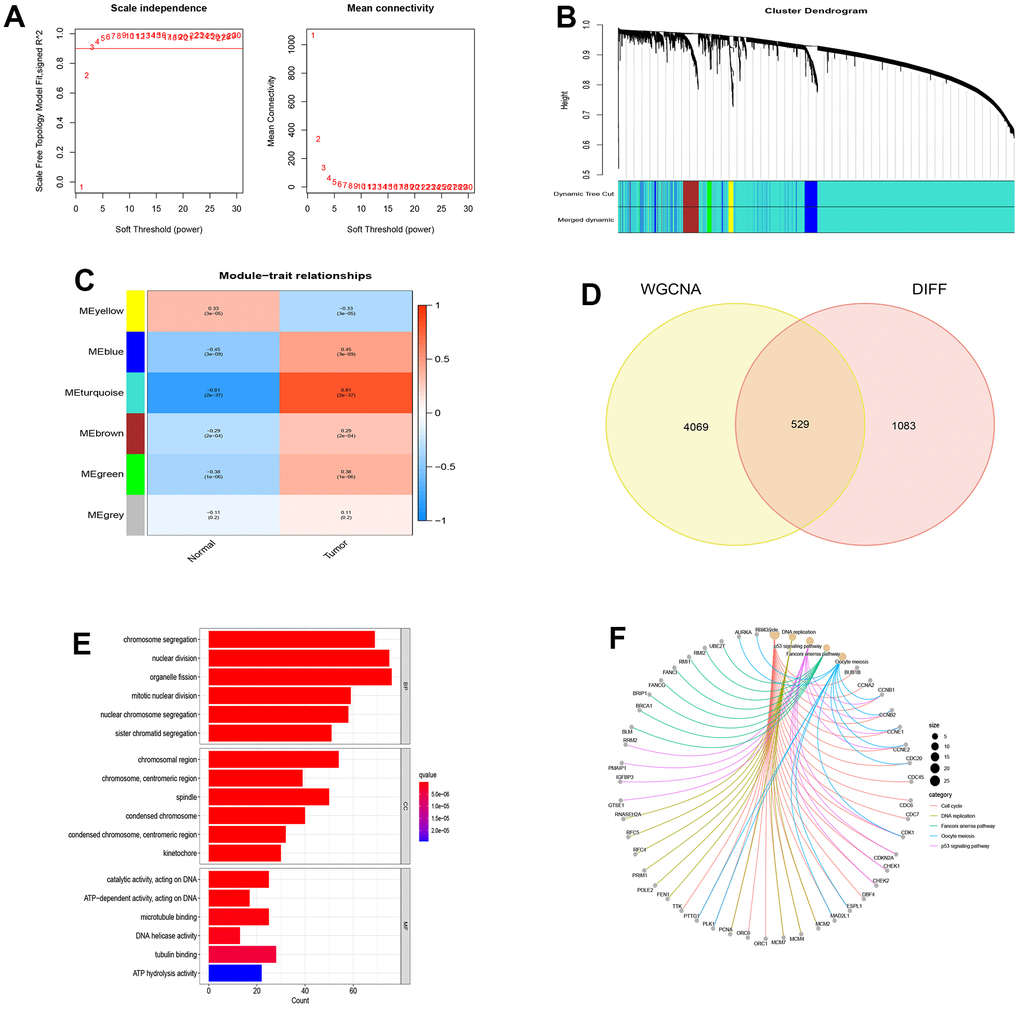
Figure 2. WGCNA analysis and module identification. (A) Scale-free exponent and average connectivity of different soft threshold powers (β); (B) Construction of gene co-expression modules; (C) Correlation analysis between different modules and tumor tissues; (D) Venn diagram of WGCNA and differentially expressed genes; (E) GO analysis of disease genes; (F) KEGG analysis of disease genes.
PPI network of drug targets and disease genes
Through the combination of drug targets and disease genes, 10 genes were found to be drug-disease genes (Figure 3A). The first PPI network was obtained based on the STRING database and Cytoscape. The subnetwork of 124 genes was obtained by the default setting of the MCODE plug-in in Cytoscape, using MCODE again and 41 hub genes were further screened out, including 4 drug-disease genes (Figure 3B). To explore the functional annotations of these hub genes, we performed DO, KEGG and GO enrichment analyses. DO analysis shows that these genes are still highly enriched for NSCLC (Figure 3C). The KEGG pathway enrichment analysis results are shown in Figure 3D. For the GO functional annotation of genes, the top 10 annotation results of BP, CC, and MF are shown in the Figure 3E. At the same time, the expression comparison between tumor tissue and normal tissue found that these hub genes were highly expressed in tumor tissue (Figure 3F). The results of ROC analysis showed that all 41 genes had good diagnostic performance for NSCLC (Supplementary Figures 1, 2).
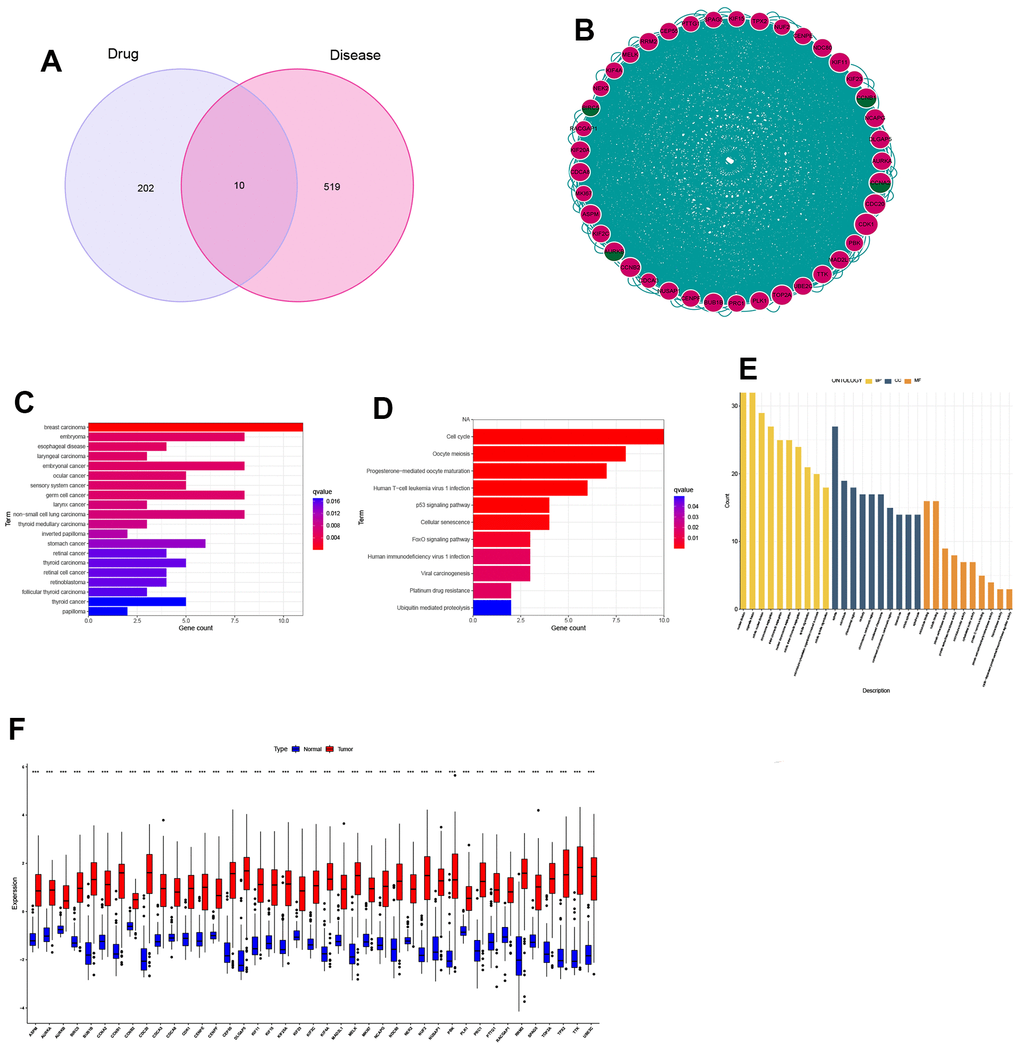
Figure 3. Identification and expression analysis of hub genes. (A) Venn diagram of drug targets and disease genes; (B) Protein-protein interaction (PPI) network of 41 hub genes; (C) DO analysis; (D) KEGG analysis; (E) GO analysis; (F) Gene expression between normal and tumor tissues.
Analysis of tumor cell immune infiltration in HNSCC
We performed immune infiltration analysis for normal and tumor groups using CIBERSORT algorithms, and the results are shown in Figure 4A. These calculation results allow us to intuitively understand that there are significantly more Plasma cells, various types of T cells, Macrophages M1 and Macrophages M2 in tumor tissues. The results of correlation analysis between 41 hub genes and immune cells are shown in Figure 4B. At the same time, we found that four drug-disease genes (CCNB1, CCNA2, BIRC5 and AURKB) were negatively correlated with monocytes, and had statistical significance, but showed different correlations with other types of immune cells (Figure 4C).
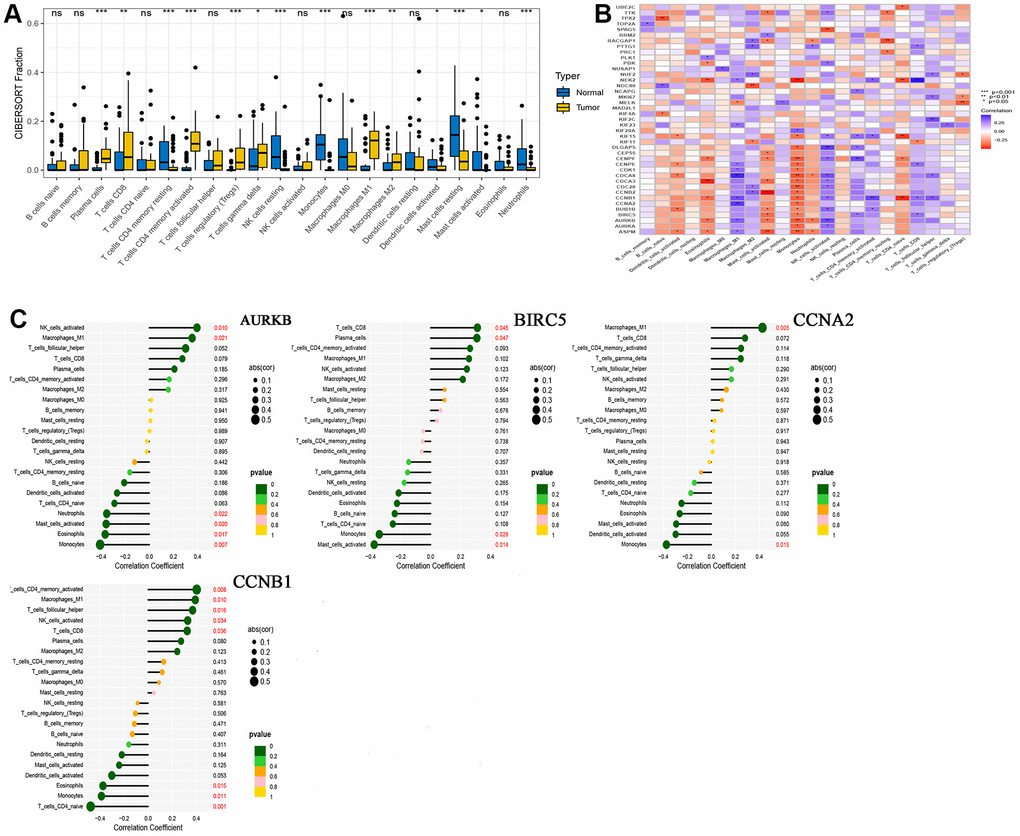
Figure 4. CIBERSORT analysis results. (A) Comparison of immune cell infiltration in normal and tumor tissues; (B) Correlation between 41 hub genes and immune cells; (C) A lollipop graph of the relationship between 4 drug-disease genes and different immune cells.
Single cell sequencing and drug-disease gene analysis
We selected the first five samples of the single-cell dataset (GSE148071), and visualized the QC metrics as a violin after following the quality control (QC) criteria of expressing more than 200 genes and less than 10,000 genes per cell, and mitochondrial content <20% (Figure 5A), while finding a positive correlation with total intracellular sequences (R = 0.91, Figure 5B), we also found a slight correlation between sequencing depth and rRNA (R = 0.08). A total of 2,000 highly variable features were identified from 27,527 features (Figure 5C). When performing PCA dimensionality reduction screening, we selected 2000 highly variable features as input, and finally selected 20 PCs with significant differences for further analysis (Figure 5D, 5E). Using the UMAP algorithm, we clustered the cells into 23 clusters and identified 15058 marker genes. In the heatmap, the top 10% of marker genes in each cluster are represented (Figure 6A). 23 clusters were annotated according to marker genes: cluster 2/10 is B cells, cluster 21 is Endothelial cells, cluster 0/4/12/14/15/20 is Epithelial cells, cluster 18 is Fibroblasts, cluster 3/5/7/16/22 is Macrophage, cluster 6/17/19 is Monocyte, cluster 8/13 is T cells, cluster 1/9/11 is Tissue stem cells (Figure 6B, 6C). The up-regulated differentially expressed genes were retained by the FindAllMarkers function and combined with the four drug-disease genes in the hub genes to analyze the distribution of these genes in cell subtypes (Figure 6D). According to the data in Figure 7A, 7B, it can be known that these four genes are highly expressed in epithelial cells, macrophages, T cells and fibroblasts.
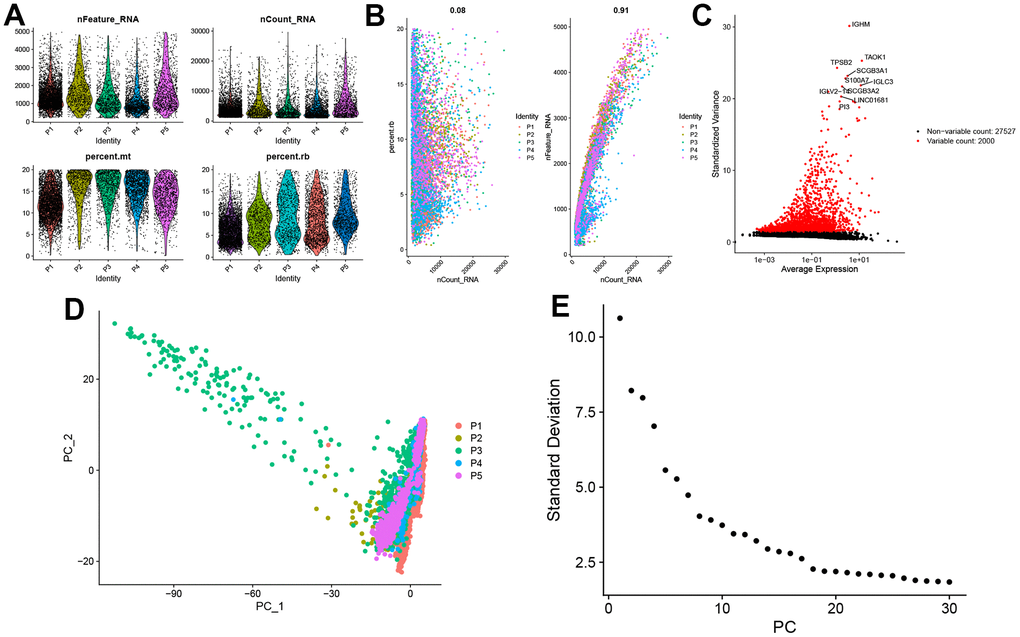
Figure 5. Single-cell sequencing data quality control and dimensionality reduction. (A) Relationship between mRNA/UMI/mitochondrial content/rRNA content of each sample after filtration. (B) The relationship between rRNA and UMI, and the relationship between mRNA and UMI; (C) The volcano map of the top 2000 hypervariable genes; (D) PCA dimensionality reduction sample distribution map; (E) PCA anchor point map.
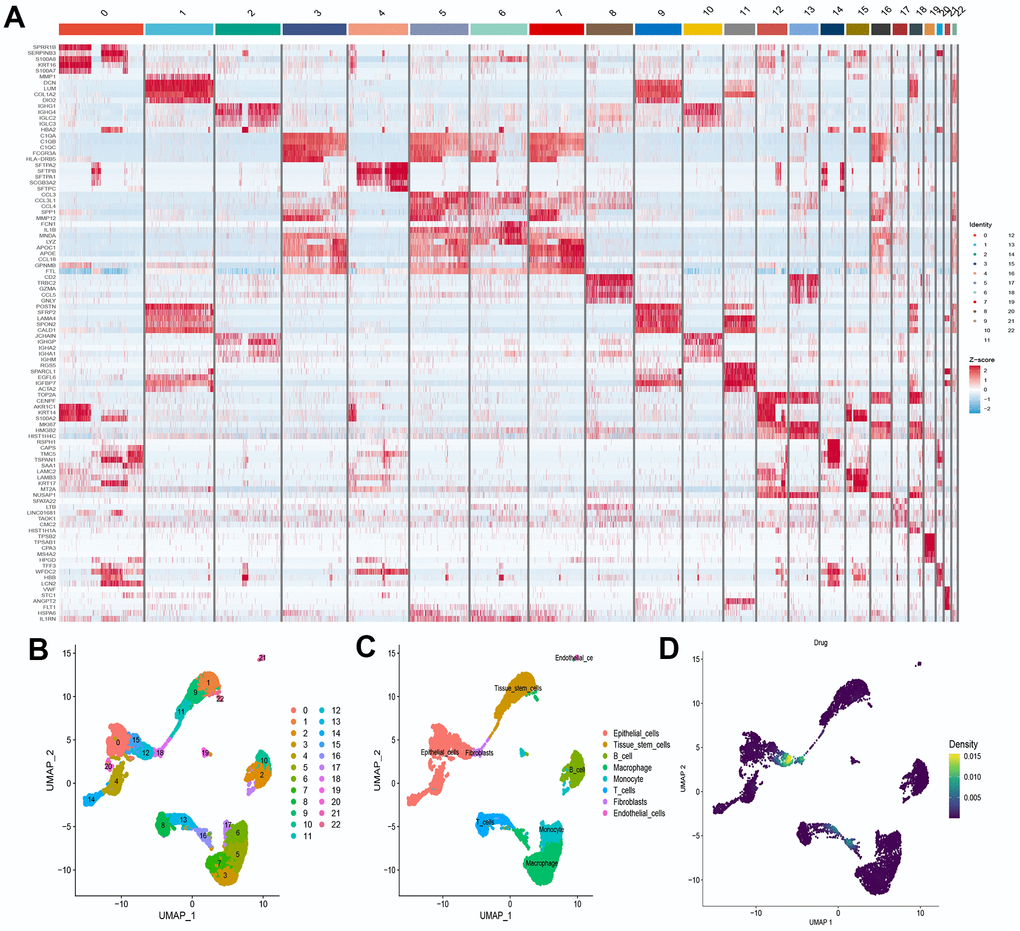
Figure 6. Definition and analysis of cell clusters. (A) Heatmap of differentially expressed genes in different cell clusters; (B) Uniform Manifold Approximation and Projection (UMAP) of 23 cell clusters; (C) The cell types were identified by marker genes; (D) Distribution of 4 drug-disease genes in different cells.
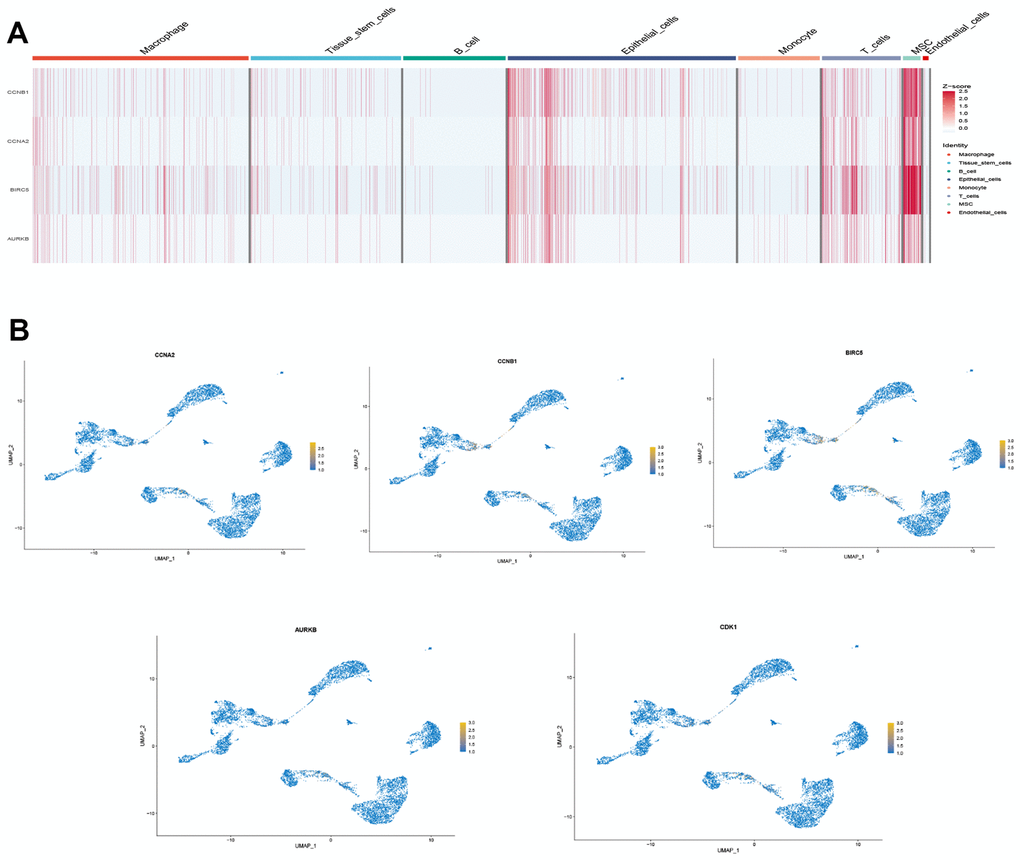
Figure 7. Drug-disease genes and single cell sequencing. (A) Heat map of expression of 4 genes among 8 types of cells; (B) Display of the distribution of each gene.
SMR analysis reveals potential key gene in NSCLC development
Based on the eQTLGen dataset, a total of 79, 87, 106, and 174 cis-eQTLs were identified for the CCNB1, CCNA2, BIRC5, and AURKB genes associated with the drug-disease relationship. The most significant cis-eQTLs were selected as genetic tools for targeting these genes. In Figure 8, the results of the SMR analysis revealed a significant increase in gene expression of CCNA2 and CCNB1 in blood samples associated with an increased risk of NSCLC occurrence. Specifically, CCNA2 showed an odds ratio (OR) of 2.024 (95% confidence interval [CI]: 1.078–3.803; P = 0.028), suggesting its potential importance in the development of NSCLC and warranting further investigation. However, although BIRC5 and AURKB showed a trend towards association with NSCLC occurrence, the corresponding P-values did not reach statistical significance. Table 1 presents the representative eQTLs for each gene and the results of the heterogeneity test (HEIDI), which indicate no significant heterogeneity among the selected eQTLs (P ≥ 0.01).
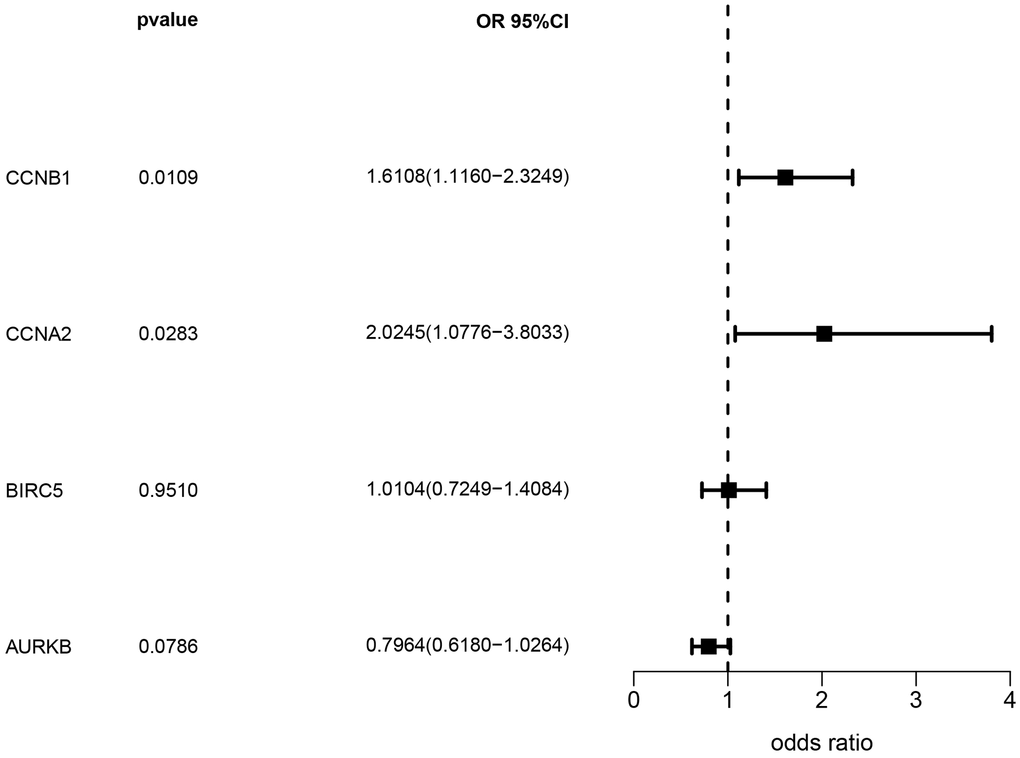
Figure 8. Analysis results of SMR.
Table 1. Representative eQTL for SMR genes and HEIDI tests.
| Gene | Chromosome location | Top eQTL | F-statistic | HEIDI (P-value) | HEIDI (Number of eQTLs) |
| CCNB1 | 5 | rs352626 | 256.70482 | 3.37E-01 | 16 |
| CCNA2 | 4 | rs4833235 | 87.343871 | 3.66E-02 | 4 |
| BIRC5 | 17 | rs11077350 | 266.91761 | 4.13E-01 | 4 |
| AURKB | 17 | rs12938531 | 246.57431 | 3.45E-01 | 20 |
Tetrandrine reduced the viability, proliferation as well as induced the cell death in NSCLC cells
In order to explore the anticancer properties of tetrandrine on NSCLC, different doses of tetrandrine (0-30 μM) were treated in the NSCLC cells. Interestingly, we found that tetrandrine had the potentiality to restrain the viability of NSCLC cells (Figure 9A, 9B). And the IC50s in A549 and H1299 cells were 9.50 μM and 10.18 μM, respectively. Therefore, 10 μM treatment of tetrandrine was selected for the next experiment. Next, we wonder whether tetrandrine treatment could suppress NSCLC cell proliferation. As expected, tetrandrine treatment caused a remarkable reduction in the percentage of EdU+ cells compared with the ctrl-group, reveling that tetrandrine significantly disrupted NSCLC growth (Figure 9C, 9D). Consequently, we utilized the Annexin V-FITC/PI assay to further clarify the potential effect of tetrandrine, and the experiment demonstrated that tetrandrine treatment significantly increased the amounts of dead cells (Figure 9E, 9F). Collectively, these results highlighted the anticancer effect of tetrandrine.
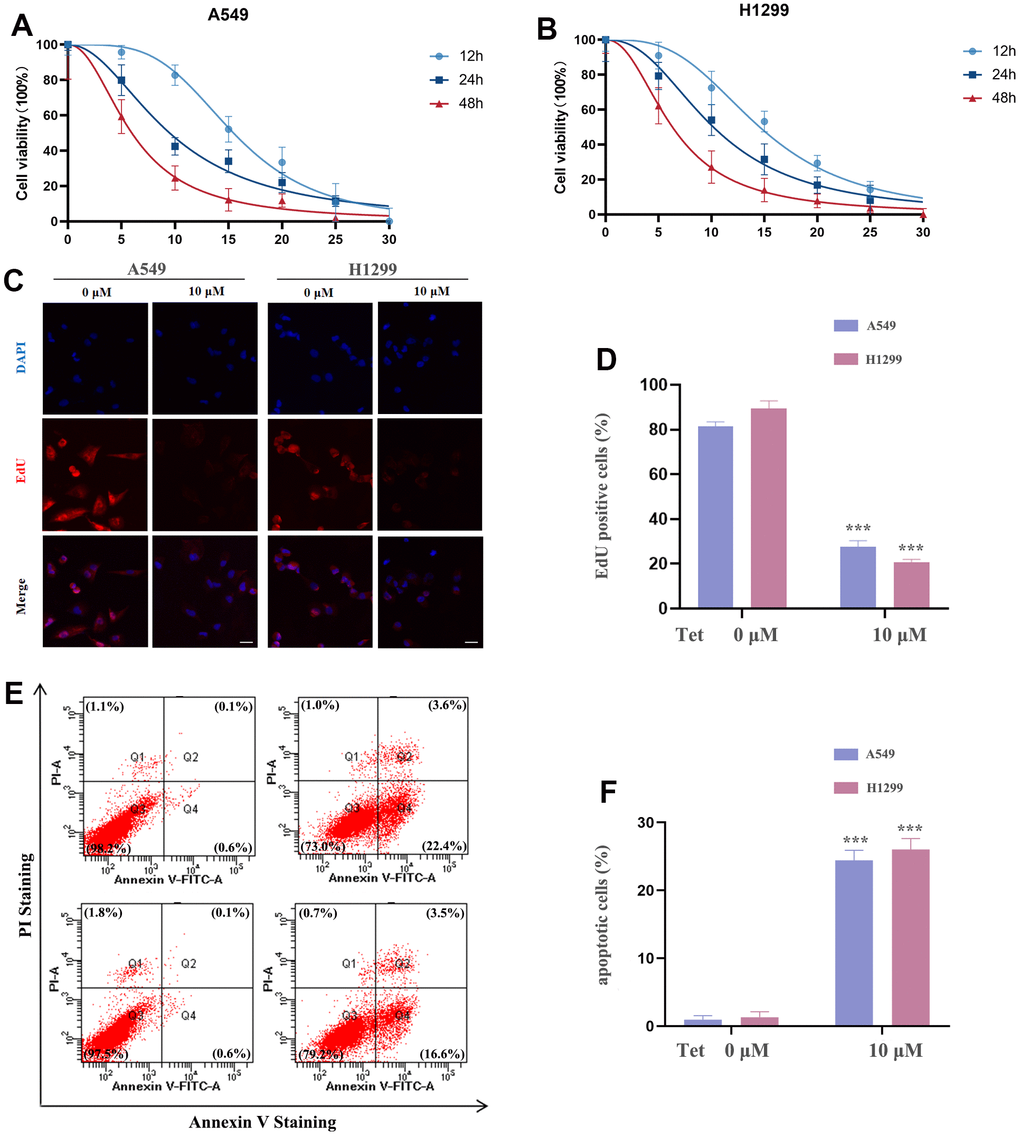
Figure 9. The effect of tetrandrine treatment on NSCLC cells. (A, B) NSCLC cells were subjected to several concentrations of tetrandrine treatment and the cell viability was measured by CCK-8 assay; (C, D) Cell proliferation of NSCLC cells were conducted by 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay; (E, F) NSCLC cells were subjected to tetrandrine treatment (10 μm, 24 h) and followed by Annexin V-FITC/PI assay. *P < 0.05, **P < 0.01, ***P < 0.001 versus control.
Molecular docking of tetrandrine with CCNA2 and CCNB1
Based on the above analysis, CCNB1, CCNA2, BIRC5, and AURKB are considered to be important genes for tetrandrine in NSCLC treatment. Single-cell sequencing results show that CCNA2 and CCNB1 are distributed in epithelial cells, and SMR shows that CCNA2 and CCNB1 are closely related to the occurrence of NSCLC. So we decided to carry out molecular docking on them. We identified 20 molecular docking patterns of tetrandrine with CCNA2 and CCNB1 (Table 2). Through the Protein-Ligand Interaction Profiler online platform, the analysis indicates that CCNA2 exhibits hydrogen bonding and hydrophobic interactions with tetrandrine (Figure 10A, 10B). In contrast, CCNB1 demonstrates hydrophobic interactions with tetrandrine (Figure 10C, 10D), consolidating the possibility of tetrandrine’s modulation through these specified genes. All procedures of this study can be reviewed in Supplementary Figure 3.
Table 2. CCNA2 and CCNB1 molecular docking results.
| Mode | Affinity(kcal/mol) (CCNA2) | Affinity(kcal/mol) (CCNB1) |
| 1 | -8.2 | -7.5 |
| 2 | -7.9 | -7.4 |
| 3 | -7.7 | -7 |
| 4 | -7.6 | -6.8 |
| 5 | -7.5 | -6.8 |
| 6 | -7.2 | -6.7 |
| 7 | -7.1 | -6.6 |
| 8 | -7.1 | -6.6 |
| 9 | -7.1 | -6.6 |
| 10 | -7.1 | -6.5 |
| 11 | -7.0 | -6.5 |
| 12 | -6.8 | -6.5 |
| 13 | -6.8 | -6.5 |
| 14 | -6.7 | -6.3 |
| 15 | -6.6 | -6.3 |
| 16 | -6.5 | -6.2 |
| 17 | -6.5 | -6.2 |
| 18 | -6.4 | -6.2 |
| 19 | -6.4 | -6 |
| 20 | -6.3 | -5.9 |
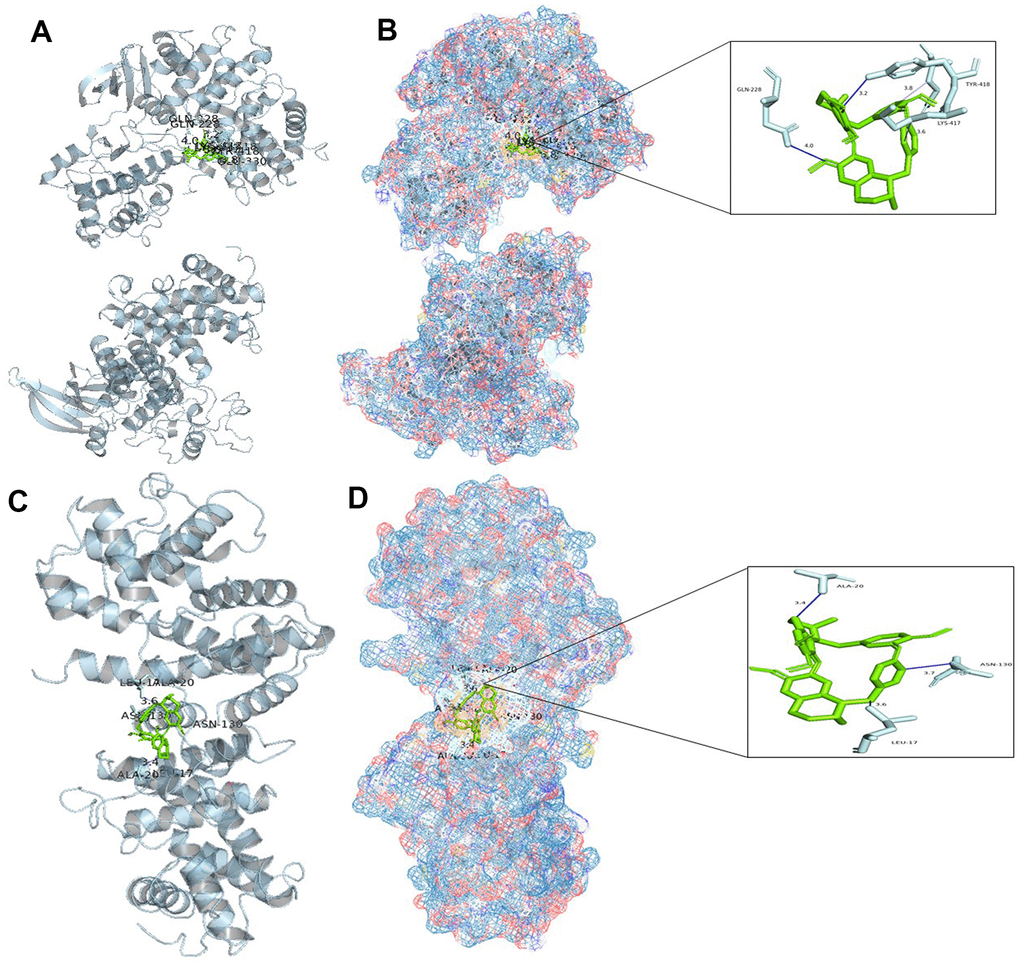
Figure 10. Molecular docking analysis. (A) Binding mode of CCNA2 protein and tetrandrine; (B) Three-dimensional (3D) interaction map of CCNA2 protein and tetrandrine; (C) Binding mode of CCNB1 protein and tetrandrine; (D) Three-dimensional (3D) interaction map of CCNB1 protein and tetrandrine.
Discussion
Although the current treatment methods for non-small cell lung cancer include surgery, radiotherapy, chemotherapy, targeted therapy, and immunotherapy, the treatment of NSCLC patients remains a serious challenge due to emerging toxic side effects and drug resistance [31–34]. Traditional Chinese medicine has a long history of treating lung cancer, and in-depth exploration of the role of traditional Chinese medicine in cancer treatment may provide us with a new breakthrough.
In a previous study, a series of novel tetrandrine derivatives were successfully synthesized via the Suzuki-Miyaura reaction and their cytotoxicity against human non-small cell lung cancer (NSCLC) A549 cells was evaluated. The results showed that compounds Y5, Y6, Y9 and Y11 had the most significant cytotoxic effects, with IC50 values ranging from 3.87 to 4.66 mM. This discovery will help in the design of more effective chemotherapy drugs for lung cancer in the future [35]. Some scholars found that inhibition of mitochondrial ATP production in lung adenocarcinoma A549 cells can enhance the cytotoxicity of tetrandrine [36]. Studies have pointed out that tetrandrine may increase the sensitivity of PC14 cells to gefitinib through lysosome inhibition [37].
We observed that tetrandrine could inhibit the viability and proliferation of NSCLC cells and induce cell death. However, the relationship between drugs and diseases is very complex. We aim to explore their relationship in various ways. 213 drug target genes were got from four drug target databases, and DO analysis confirmed that these drug targets mainly play a role in various cancers including NSCLC. KEGG analysis confirmed that these genes were enriched in EGFR tyrosine kinase inhibitor resistance [38], PI3K-Akt signaling pathway [39], PD-L1 expression and PD-1 checkpoint pathway [40] and other pathways. GO analysis also confirmed that drug genes were enriched in peptide-serine modification, synaptic membrane and protein serine/threonine kinase activity. Many studies have confirmed that the above enrichment results are closely related to NSCLC [41, 42]. Then we adopted the method of WGCNA interacting with differentially expressed genes. Finally, we obtained 529 disease genes.
Using the interactions between drug targets and disease genes, we identified 10 drug-disease genes and constructed a PPI network covering all drug and disease genes. Through two rounds of MCODE screening, we screened out a core gene network containing four drug-disease genes. We performed DO/GO/KEGG functional enrichment analysis on these core genes and found that these genes were strongly associated with the progression of NSCLC. In particular, KEGG analysis results showed that these core genes were enriched in pathways such as platinum resistance [43], Cellular senescence [44] and P53 signaling pathway [45], This also suggests that the hub gene may affect NSCLC through those ways.
After immune infiltration analysis, we found that the number of various types of T cells and macrophages increased in tumor tissues. On the one hand, it shows that the antigens expressed by tumor cells can be recognized by the immune system, thereby activating T cells and macrophages. On the other hand, tumor cells secrete a variety of growth factors and cytokines, such as interleukin 6 (IL-6), tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ), which may also lead to such results [46, 47]. At the same time, we found that four drug-disease genes (CCNB1, CCNA2, BIRC5, and AURKB) were highly expressed in tumors, with high diagnostic accuracy, and were positively correlated with plasma cells, T cells, and macrophages; while negatively correlated with monocytes. Studies have shown that T cells and B cells play an important role in tumor immunity [48, 49]. If these drug-disease genes can be used as new biomarkers, it may provide new ideas for current immunotherapy.
Single-cell sequencing results showed that multiple cell types still exist in NSCLC at the cellular level. In addition, four genes showed higher expression in epithelial and endothelial cells compared to other cell types. Combined with previous bioinformatics data, suggest that they may be involved in specific biological processes in epithelial and endothelial cells, such as cell proliferation, metastasis and angiogenesis, which play a crucial role in the development and progression of NSCLC. To overcome the limitations of traditional observational studies, the SMR approach was used in this study, utilizing genetic variants associated with the expression of four genes as instrumental variables. The analysis showed that increased expression of CCNA2 and CCNB1 was associated with an increased risk of NSCLC formation. Research has revealed a significant upregulation of CCNA2 mRNA and lncRNA DNAH17 antisense RNA 1 (DNAH17-AS1) in NSCLC specimens and cell lines, accompanied by a notable decrease in miR-877-5p expression. Elevated DNAH17-AS1 levels correlate with TNM staging, distant metastasis, and reduced overall survival. Functional analysis indicates that silencing DNAH17-AS1 inhibits proliferation, migration, and invasion of H1299 and 95D cells while promoting apoptosis. Further mechanistic investigations suggest that DNAH17-AS1 may function as a miR-877-5p sponge to upregulate CCNA2, thereby exerting its oncogenic role [50]. Simultaneously, research has also shown that Danshensu IIA can inhibit the progression of lung adenocarcinoma by inducing apoptosis and cell cycle arrest. These effects are achieved through the regulation of the CCNA2-CDK2 complex and the AURKA/PLK1 pathway [51]. Previous studies have indicated that MEOX1 exhibits lower expression in lung cancer tissues compared to normal adjacent tissues. MEOX1 shows a positive correlation with the overall survival of lung cancer patients, particularly those with lung adenocarcinoma. In vitro and in vivo functional experiments have demonstrated that stable overexpression of MEOX1 significantly inhibits the proliferative capacity of NSCLC cells, induces cell cycle arrest in the G2 phase, and enhances apoptotic capabilities. Furthermore, researchers have found a negative correlation between MEOX1 and CCNB1 mRNA expression in various lung cancer tissues. Further investigations have revealed that MEOX1 suppresses the progression of lung cancer cells by inhibiting the cell cycle checkpoint gene CCNB1 [52]. Bao et al. [53] discovered that overexpression of CCNB1 promotes the progression of lung cancer cells, and miR-139-5p may act as a negative regulator of CCNB1, thereby inhibiting cell proliferation, migration, invasion, and cell cycle progression. Our molecular docking studies of CCNA2 and CCNB1 with tetrandrine have revealed interaction sites, highlighting the significant potential of tetrandrine in the treatment of NSCLC. This presents a crucial opportunity for repurposing as compared to the development of new drugs.
Conclusions
In this study, we systematically investigated the relationship between tetrandrine and non-small cell lung cancer (NSCLC), confirming the relevance and importance of drug-disease genes in tumor immunity, cell subtypes, and disease progression. It is also well illustrated that the repurposing of tetrandrine could potentially contribute to the treatment of NSCLC.
Abbreviations
BP: biological process;
CHM: Chinese herbal medicine;
CNB: Cinobufacin;
CTD: Comparative Toxicogenomics Database;
CC: cellular component;
DO: Disease Ontology;
GO: Gene Ontology;
ICAM-1: intercellular adhesion molecule-1;
KEGG: Kyoto Encyclopedia of Genes and Genomes;
MF: molecular function;
NSCLC: Non-small cell lung cancer;
PPI: protein-protein interaction;
PCA: Principal Component Analysis;
QC: quality control;
ROC: receiver operating characteristic;
TB: theabrownin;
UMAP: Uniform Manifold Approximation and Projection;
VEGF: vascular endothelial growth factor;
YYJD: Yiqi Yangyin Jiedu Decoction.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by JHL, XCM. Vitro cell experiments were performed by DH. The first draft of the manuscript was written by JHL, XCM and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We want to acknowledge the participants and investigators of the FinnGen study.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Ethical Statement and Consent
This study is based on publicly available summary-level data from genome-wide association studies (GWAS) and expression quantitative trait loci (eQTL) studies. These studies have been approved by relevant institutional review boards and informed consent has been obtained from the participants.
Funding
This study was supported by the National Natural Science Foundation of China (no. 81774376), the Science and Technology Foundation of Guangzhou (no. 201803010059), the National Natural Science Foundation of China (no.81900160) and the Science and Technology Planning Project of Guangzhou under Grant 2023A04J1919.
References
-
1.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021; 71:209–49. https://doi.org/10.3322/caac.21660 [PubMed]
-
2.
Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021; 398:535–54. https://doi.org/10.1016/S0140-6736(21)00312-3 [PubMed]
-
3.
Jasper K, Stiles B, McDonald F, Palma DA. Practical Management of Oligometastatic Non-Small-Cell Lung Cancer. J Clin Oncol. 2022; 40:635–41. https://doi.org/10.1200/JCO.21.01719 [PubMed]
-
4.
Chen P, Liu Y, Wen Y, Zhou C. Non-small cell lung cancer in China. Cancer Commun (Lond). 2022; 42:937–70. https://doi.org/10.1002/cac2.12359 [PubMed]
-
5.
Tian X, Gu T, Lee MH, Dong Z. Challenge and countermeasures for EGFR targeted therapy in non-small cell lung cancer. Biochim Biophys Acta Rev Cancer. 2022; 1877:188645. https://doi.org/10.1016/j.bbcan.2021.188645 [PubMed]
-
6.
Luo YH, Chiu CH, Scott Kuo CH, Chou TY, Yeh YC, Hsu HS, Yen SH, Wu YH, Yang JC, Liao BC, Hsia TC, Chen YM. Lung Cancer in Republic of China. J Thorac Oncol. 2021; 16:519–27. https://doi.org/10.1016/j.jtho.2020.10.155 [PubMed]
-
7.
Jiao L, Xu J, Sun J, Chen Z, Gong Y, Bi L, Lu Y, Yao J, Zhu W, Hou A, Feng G, Jia Y, Shen W, et al. Chinese Herbal Medicine Combined With EGFR-TKI in EGFR Mutation-Positive Advanced Pulmonary Adenocarcinoma (CATLA): A Multicenter, Randomized, Double-Blind, Placebo-Controlled Trial. Front Pharmacol. 2019; 10:732. https://doi.org/10.3389/fphar.2019.00732 [PubMed]
-
8.
Xiao X, Guo L, Dai W, Yan B, Zhang J, Yuan Q, Zhou L, Shan L, Efferth T. Green tea-derived theabrownin suppresses human non-small cell lung carcinoma in xenograft model through activation of not only p53 signaling but also MAPK/JNK signaling pathway. J Ethnopharmacol. 2022; 291:115167. https://doi.org/10.1016/j.jep.2022.115167 [PubMed]
-
9.
Zhang L, Liang B, Xu H, Gong Y, Hu W, Jin Z, Wu X, Chen X, Li M, Shi L, Shi Y, Wang Y, Yang L. Cinobufagin induces FOXO1-regulated apoptosis, proliferation, migration, and invasion by inhibiting G9a in non-small-cell lung cancer A549 cells. J Ethnopharmacol. 2022; 291:115095. https://doi.org/10.1016/j.jep.2022.115095 [PubMed]
-
10.
Chu S, Lu Y, Liu W, Ma X, Peng J, Wang X, Jiang M, Bai G. Ursolic acid alleviates tetrandrine-induced hepatotoxicity by competitively binding to the substrate-binding site of glutathione S-transferases. Phytomedicine. 2022; 104:154325. https://doi.org/10.1016/j.phymed.2022.154325 [PubMed]
-
11.
Cho HS, Chang SH, Chung YS, Shin JY, Park SJ, Lee ES, Hwang SK, Kwon JT, Tehrani AM, Woo M, Noh MS, Hanifah H, Jin H, et al. Synergistic effect of ERK inhibition on tetrandrine-induced apoptosis in A549 human lung carcinoma cells. J Vet Sci. 2009; 10:23–8. https://doi.org/10.4142/jvs.2009.10.1.23 [PubMed]
-
12.
Qiu W, Su M, Xie F, Ai J, Ren Y, Zhang J, Guan R, He W, Gong Y, Guo Y. Tetrandrine blocks autophagic flux and induces apoptosis via energetic impairment in cancer cells. Cell Death Dis. 2014; 5:e1123. https://doi.org/10.1038/cddis.2014.84 [PubMed]
-
13.
Zhou Y, Mu L, Liu XL, Li Q, Ding LX, Chen HC, Hu Y, Li FS, Sun WJ, He BC, Wu K. Tetrandrine inhibits proliferation of colon cancer cells by BMP9/ PTEN/ PI3K/AKT signaling. Genes Dis. 2019; 8:373–83. https://doi.org/10.1016/j.gendis.2019.10.017 [PubMed]
-
14.
Chen Z, Zhao L, Zhao F, Yang G, Wang JJ. Tetrandrine suppresses lung cancer growth and induces apoptosis, potentially via the VEGF/HIF-1α/ICAM-1 signaling pathway. Oncol Lett. 2018; 15:7433–7. https://doi.org/10.3892/ol.2018.8190 [PubMed]
-
15.
He Z, Gao K, Dong L, Liu L, Qu X, Zou Z, Wu Y, Bu D, Guo JC, Zhao Y. Drug screening and biomarker gene investigation in cancer therapy through the human transcriptional regulatory network. Comput Struct Biotechnol J. 2023; 21:1557–72. https://doi.org/10.1016/j.csbj.2023.02.005 [PubMed]
-
16.
Davis AP, Wiegers TC, Johnson RJ, Sciaky D, Wiegers J, Mattingly CJ. Comparative Toxicogenomics Database (CTD): update 2023. Nucleic Acids Res. 2023; 51:D1257–62. https://doi.org/10.1093/nar/gkac833 [PubMed]
-
17.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019; 47:W357–64. https://doi.org/10.1093/nar/gkz382 [PubMed]
-
18.
Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016; 44:D1045–53. https://doi.org/10.1093/nar/gkv1072 [PubMed]
-
19.
Yao ZJ, Dong J, Che YJ, Zhu MF, Wen M, Wang NN, Wang S, Lu AP, Cao DS. TargetNet: a web service for predicting potential drug-target interaction profiling via multi-target SAR models. J Comput Aided Mol Des. 2016; 30:413–24. https://doi.org/10.1007/s10822-016-9915-2 [PubMed]
-
20.
Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010; 5:e10312. https://doi.org/10.1371/journal.pone.0010312 [PubMed]
-
21.
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021; 49:D605–12. https://doi.org/10.1093/nar/gkaa1074 [PubMed]
-
22.
Wu F, Fan J, He Y, Xiong A, Yu J, Li Y, Zhang Y, Zhao W, Zhou F, Li W, Zhang J, Zhang X, Qiao M, et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun. 2021; 12:2540. https://doi.org/10.1038/s41467-021-22801-0 [PubMed]
-
23.
Võsa U, Claringbould A, Westra HJ, Bonder MJ, Deelen P, Zeng B, Kirsten H, Saha A, Kreuzhuber R, Yazar S, Brugge H, Oelen R, de Vries DH, et al, and BIOS Consortium, and i2QTL Consortium. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021; 53:1300–10. https://doi.org/10.1038/s41588-021-00913-z [PubMed]
-
24.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, Loukola A, Lahtela E, Mattsson H, et al, and FinnGen. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023; 613:508–18. https://doi.org/10.1038/s41586-022-05473-8 [PubMed]
-
25.
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016; 48:481–7. https://doi.org/10.1038/ng.3538 [PubMed]
-
26.
Chauquet S, Zhu Z, O’Donovan MC, Walters JTR, Wray NR, Shah S. Association of Antihypertensive Drug Target Genes With Psychiatric Disorders: A Mendelian Randomization Study. JAMA Psychiatry. 2021; 78:623–31. https://doi.org/10.1001/jamapsychiatry.2021.0005 [PubMed]
-
27.
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–95. https://doi.org/10.1093/nar/gkaa971 [PubMed]
-
28.
Goodsell DS, Zardecki C, Di Costanzo L, Duarte JM, Hudson BP, Persikova I, Segura J, Shao C, Voigt M, Westbrook JD, Young JY, Burley SK. RCSB Protein Data Bank: Enabling biomedical research and drug discovery. Protein Sci. 2020; 29:52–65. https://doi.org/10.1002/pro.3730 [PubMed]
-
29.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010; 31:455–61. https://doi.org/10.1002/jcc.21334 [PubMed]
-
30.
Adasme MF, Linnemann KL, Bolz SN, Kaiser F, Salentin S, Haupt VJ, Schroeder M. PLIP 2021: expanding the scope of the protein-ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021; 49:W530–4. https://doi.org/10.1093/nar/gkab294 [PubMed]
-
31.
Reck M, Remon J, Hellmann MD. First-Line Immunotherapy for Non-Small-Cell Lung Cancer. J Clin Oncol. 2022; 40:586–97. https://doi.org/10.1200/JCO.21.01497 [PubMed]
-
32.
Tan AC, Tan DSW. Targeted Therapies for Lung Cancer Patients With Oncogenic Driver Molecular Alterations. J Clin Oncol. 2022; 40:611–25. https://doi.org/10.1200/JCO.21.01626 [PubMed]
-
33.
Abuhelwa Z, Alloghbi A, Nagasaka M. A comprehensive review on antibody-drug conjugates (ADCs) in the treatment landscape of non-small cell lung cancer (NSCLC). Cancer Treat Rev. 2022; 106:102393. https://doi.org/10.1016/j.ctrv.2022.102393 [PubMed]
-
34.
Mithoowani H, Febbraro M. Non-Small-Cell Lung Cancer in 2022: A Review for General Practitioners in Oncology. Curr Oncol. 2022; 29:1828–39. https://doi.org/10.3390/curroncol29030150 [PubMed]
-
35.
Yang QH, Jiang CS, Jin T, Xu JF, Qu TL, Guo YW, Zhao ZB. Synthesis of novel tetrandrine derivatives and their inhibition against NSCLC A549 cells. J Asian Nat Prod Res. 2018; 20:1064–74. https://doi.org/10.1080/10286020.2018.1478817 [PubMed]
-
36.
Chow LWC, Cheng KS, Leong F, Cheung CW, Shiao LR, Leung YM, Wong KL. Enhancing tetrandrine cytotoxicity in human lung carcinoma A549 cells by suppressing mitochondrial ATP production. Naunyn Schmiedebergs Arch Pharmacol. 2019; 392:427–36. https://doi.org/10.1007/s00210-018-01601-2 [PubMed]
-
37.
Sato E, Ohta S, Kawakami K, Ikeda M, Takahashi T, Kobayashi S, Nomura M. Tetrandrine Increases the Sensitivity of Human Lung Adenocarcinoma PC14 Cells to Gefitinib by Lysosomal Inhibition. Anticancer Res. 2019; 39:6585–93. https://doi.org/10.21873/anticanres.13874 [PubMed]
-
38.
Cooper AJ, Sequist LV, Lin JJ. Third-generation EGFR and ALK inhibitors: mechanisms of resistance and management. Nat Rev Clin Oncol. 2022; 19:499–514. https://doi.org/10.1038/s41571-022-00639-9 [PubMed]
-
39.
Liu Z, Huang M, Hong Y, Wang S, Xu Y, Zhong C, Zhang J, Zhuang Z, Shan S, Ren T. Isovalerylspiramycin I suppresses non-small cell lung carcinoma growth through ROS-mediated inhibition of PI3K/AKT signaling pathway. Int J Biol Sci. 2022; 18:3714–30. https://doi.org/10.7150/ijbs.69989 [PubMed]
-
40.
Liu C, Zheng S, Wang Z, Wang S, Wang X, Yang L, Xu H, Cao Z, Feng X, Xue Q, Wang Y, Sun N, He J. KRAS-G12D mutation drives immune suppression and the primary resistance of anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer. Cancer Commun (Lond). 2022; 42:828–47. https://doi.org/10.1002/cac2.12327 [PubMed]
-
41.
He Y, Jiang X, Duan L, Xiong Q, Yuan Y, Liu P, Jiang L, Shen Q, Zhao S, Yang C, Chen Y. LncRNA PKMYT1AR promotes cancer stem cell maintenance in non-small cell lung cancer via activating Wnt signaling pathway. Mol Cancer. 2021; 20:156. https://doi.org/10.1186/s12943-021-01469-6 [PubMed]
-
42.
Lu L, Zhang X, Zhang B, Wu J, Zhang X. Synaptic acetylcholinesterase targeted by microRNA-212 functions as a tumor suppressor in non-small cell lung cancer. Int J Biochem Cell Biol. 2013; 45:2530–40. https://doi.org/10.1016/j.biocel.2013.08.007 [PubMed]
-
43.
Sui Q, Chen Z, Hu Z, Huang Y, Liang J, Bi G, Bian Y, Zhao M, Zhan C, Lin Z, Wang Q, Tan L. Cisplatin resistance-related multi-omics differences and the establishment of machine learning models. J Transl Med. 2022; 20:171. https://doi.org/10.1186/s12967-022-03372-0 [PubMed]
-
44.
Jiao D, Zheng X, Du X, Wang D, Hu Z, Sun R, Tian Z, Fu B, Wei H. Immunogenic senescence sensitizes lung cancer to LUNX-targeting therapy. Cancer Immunol Immunother. 2022; 71:1403–17. https://doi.org/10.1007/s00262-021-03077-1 [PubMed]
-
45.
Shen W, Tong D, Chen J, Li H, Hu Z, Xu S, He S, Ge Z, Zhang J, Mao Q, Chen H, Xu G. Silencing oncogene cell division cycle associated 5 induces apoptosis and G1 phase arrest of non-small cell lung cancer cells via p53-p21 signaling pathway. J Clin Lab Anal. 2022; 36:e24396. https://doi.org/10.1002/jcla.24396 [PubMed]
-
46.
Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, Gao R, Zhang L, Dong M, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018; 24:978–85. https://doi.org/10.1038/s41591-018-0045-3 [PubMed]
-
47.
Casanova-Acebes M, Dalla E, Leader AM, LeBerichel J, Nikolic J, Morales BM, Brown M, Chang C, Troncoso L, Chen ST, Sastre-Perona A, Park MD, Tabachnikova A, et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature. 2021; 595:578–84. https://doi.org/10.1038/s41586-021-03651-8 [PubMed]
-
48.
Wang DR, Wu XL, Sun YL. Therapeutic targets and biomarkers of tumor immunotherapy: response versus non-response. Signal Transduct Target Ther. 2022; 7:331. https://doi.org/10.1038/s41392-022-01136-2 [PubMed]
-
49.
Sivori S, Pende D, Quatrini L, Pietra G, Della Chiesa M, Vacca P, Tumino N, Moretta F, Mingari MC, Locatelli F, Moretta L. NK cells and ILCs in tumor immunotherapy. Mol Aspects Med. 2021; 80:100870. https://doi.org/10.1016/j.mam.2020.100870 [PubMed]
-
50.
Du LJ, Mao LJ, Jing RJ. Long noncoding RNA DNAH17-AS1 promotes tumorigenesis and metastasis of non-small cell lung cancer via regulating miR-877-5p/CCNA2 pathway. Biochem Biophys Res Commun. 2020; 533:565–72. https://doi.org/10.1016/j.bbrc.2020.09.047 [PubMed]
-
51.
Li Z, Zhang Y, Zhou Y, Wang F, Yin C, Ding L, Zhang S. Tanshinone IIA suppresses the progression of lung adenocarcinoma through regulating CCNA2-CDK2 complex and AURKA/PLK1 pathway. Sci Rep. 2021; 11:23681. https://doi.org/10.1038/s41598-021-03166-2 [PubMed]
-
52.
Xiao X, Rui B, Rui H, Ju M, Hongtao L. MEOX1 suppresses the progression of lung cancer cells by inhibiting the cell-cycle checkpoint gene CCNB1. Environ Toxicol. 2022; 37:504–13. https://doi.org/10.1002/tox.23416 [PubMed]
-
53.
Bao B, Yu X, Zheng W. MiR-139-5p Targeting CCNB1 Modulates Proliferation, Migration, Invasion and Cell Cycle in Lung Adenocarcinoma. Mol Biotechnol. 2022; 64:852–60. https://doi.org/10.1007/s12033-022-00465-5 [PubMed]