SIRT1 is localized in the nucleus of cardiomyocytes, but not of non cardiac cell types
Subcellular localization of chromatin remodeling SIRT1 varies in different cell types, and it is crucial for its impact on cellular functions. Previous reports based mainly on biochemical fractionation techniques indicated that SIRT1 is localized in the cytoplasm of cardiomyocytes in basal conditions, and nucleo-cytoplasmic shuttling is a regulatory mechanism of SIRT1 that may participate in the stress response [21, 22]. While the cardiomyocytes constitute the bulk of heart mass, this organ is composed only for ~60% of this cell type, and fibroblasts and other cell types (endothelial cells, macrophages, stem cells) account for the remaining fraction [23]. A bidirectional cross-talk between cardiomyocytes and non-cardiac cells is crucial to respond to pathological stimuli [23], and SIRT1-dependent metabolic effects may be cell- and compartment specific. To date, a quantitative microscopy-based approach to elucidate cell-specific SIRT1 localization in the heart has not been undertaken. Aiming to this, we first evaluated SIRT1 localization in the cardiomyocytes. Confocal analyses of heart cryo-sections from unstimulated 3 months old wild type (WT) and mIGF-1 transgenic (Tg) mice were performed. Sections were co-stained with SIRT1 (Fig.1 Panel A and C, green), and a specific marker of cardiomyocyte sarcomeric myosin (MF-20, Fig.1 Panel A and C, red), and DAPI counterstaining was used for the nuclei (Fig.1 Panel A and C, blue). Software-assisted analysis showed that SIRT1 was localized in ~22 % of the cardiomyocytes nuclei in WT mice and in ~28 % of the cardiomyocytes nuclei in mIGF-1 Tg mice (Fig. 1A and B). Co-staining for SIRT1 and vimentin (class III intermediate filament, marker of fibroblasts and mesenchymal like cells) revealed an exclusive cytoplasmic co-localization of SIRT1 in these non cardiac cells both in WT and mIGF-1 mice (Fig. 1, Panel C). Typically, larger SIRT1 positive nuclei in vimentin negative cells (Fig. 1 Panel C, arrowheads), presumably belonging to cardiomyocytes, were found adjacent to vimentin positive cells (Fig. 1 Panel C, arrows). SIRT1 was found exclusively in the cytoplasm also in endothelial cells, as determined by co-localization with CD144 (vascular endothelial-cadherin) (data not shown). These findings suggest that, in the heart of both WT and mIGF-1 mice, SIRT1 localizes in the nucleus of a substantial fraction of cardiomyocytes, whereas it is only cytoplasmic in non cardiac cell types.
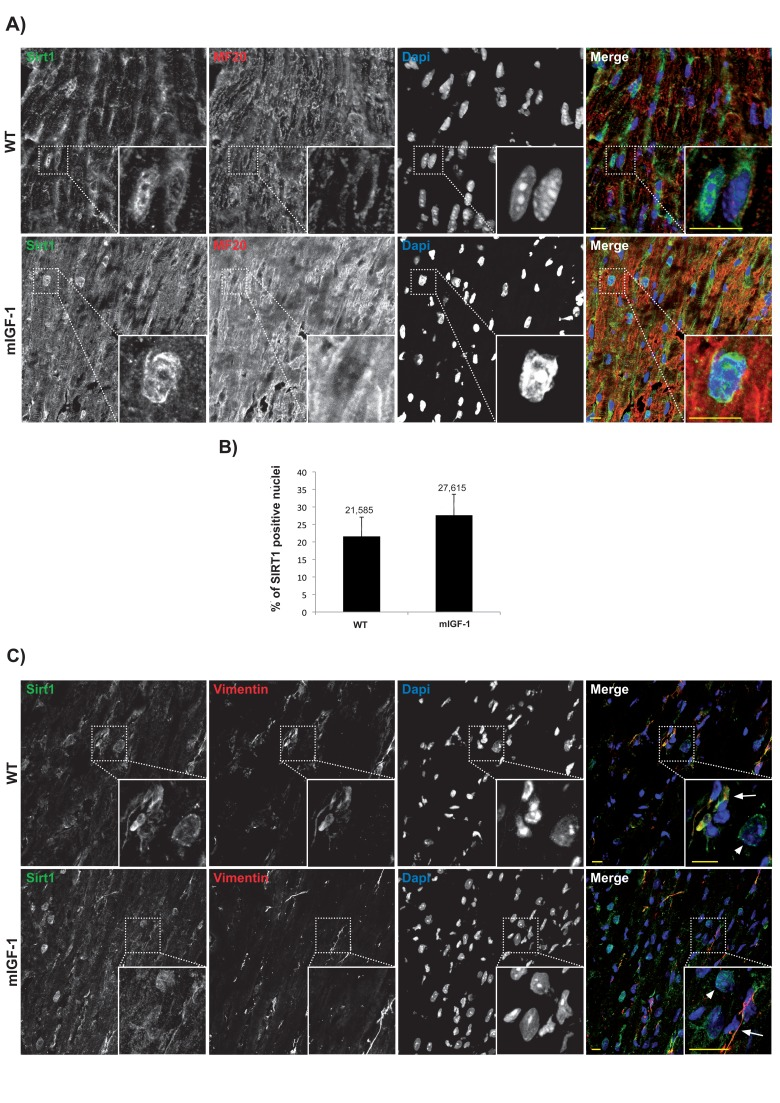
Figure 1. Confocal analysis of SIRT1 localization in the heart of wild type and mIGF-1 transgenic mice. (A) representative heart sections of a 3 months old wild type (WT, upper panel) and of a cardiac restricted mIGF-1 transgenic mouse (mIGF-1, lower panel), stained for cardiomyocyte specific marker sarcomeric myosin (red), SIRT1 (green). Nuclei are counterstained with DAPI (blue). (B) quantification of SIRT1 positive nuclei, as shown in A. Total DAPI positive nuclei considered for quantification of SIRT1 immunopositivity in WT = 2280, and in mIGF-1 Tg = 2053, in 4 independent preparations. (C) representative heart sections of a 3 months old WT (upper panel) and of a cardiac restricted mIGF-1 Tg mouse (lower panel), stained for vimentin (red), SIRT1 (green). Nuclei are counterstained with DAPI (blue). Mesenchymal cells cytoplasm (arrows) appeared positive for both vimentin (red) and SIRT1 (green). Whereas in other cell types, presumably cardiomyocytes due to nuclear localization of SIRT1 (green), vimentin is lacking in thecytoplasm (arrowheads). Scale Bars= 10 μm.
High throughput sequencing analyses of cardiac mIGF-1/SIRT1 signaling
Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) methodology [24] has been used to show that SIRT1 binds to highly repetitive genomic DNA and to a functionally diverse subset of genes in embryonic stem cells [20]. To our knowledge this is the only data set available on SIRT1 genome-wide binding. In view of the fact that we found SIRT1 localized in 22 to 28% of cardiomyocytes nuclei, with the aim of understanding the nuclear genomic function of this deacetylase, we performed genome-wide profiling of its binding to DNA in the hearts of WT or mIGF-1 Tg mice by using ChIP-Seq and a specific ChIP-grade commercial antibody. The experiments were run in duplicate for both genotypes and between 78 to 95% of the reads were mapped. The majority of SIRT1 counts localized to repetitive DNA regions (data not shown) in agreement with previous studies [20]. We selected those localized within 10 Kb from gene start site (TSS) (from www.ensembl.org). 302 SIRT1 bound TSS were identified, corresponding to 183 functional genes, 68 pseudogenes and 61 predicted genes. As shown in Fig. 2 by a common dispersion plot, while the majority of the DNA SIRT-1 bound genes were the same between WT and mIGF-1 Tg mice, a subset of genes were differentially bound by SIRT1 in the heart of either WT or mIGF-1 Tg mice. Clustering these genes according to biological functions, SIRT1 differentially bound TSS of established key players of cardiovascular function/blood pressure (ABCG1, C3, CTGF, MYH11, PDGF, TRPA1 in WT hearts; and ADRB1, ERK1/2, G protein alpha, GPCR, p38 MAPK, VEGFB, NF-kB in mIGF-1 Tg hearts), of atherosclerosis/inflammation/oxidative stress (CDKN2A, ABCG5 ABCG8, CYP7A1, ERO1LB, ERP44, FZD-1, HNF4alpha, LXR ligand RA, Nrh1, C-ERBA in WT hearts; and APOC3, ERK1/2, MEFV, NLRP2, p38 MAPK, S1PR, S1PR4, S1P, GDF15, NF-kB in mIGF-1 Tg hearts), of immunity (C3, XBP1 in WT hearts; DHX58, ERK1/2, G protein alpha, GPCR, GPR27, HLA-A, IFNA1/IFNA13, p38 MAPK, PYCARD, S1P, HLA-abc, IFI30, NF-kB in mIGF-1 Tg hearts), of behavior (IMPA1, KLF-9, PLC, TRPA1 in WT hearts; and ADR2A, ERK1/2, G protein alpha, GPCR, GPR27, NPBWR1, p38 MAPK, NF-kB in mIGF-1 Tg hearts) and of other basic and pathological functions (ABCB11, CALB1, FANCA, FANCL, FZD-1, MYB-2, MYH9, PEX-1, SDF2L1, SLC26A5, SPAM1, SYNE, THRA in WT hearts; and CHRM3, CHRM4, CTAG1B, HIST4H4, PARP10, XIST, Histone H3, KDM3A in mIGF-1 Tg hearts).
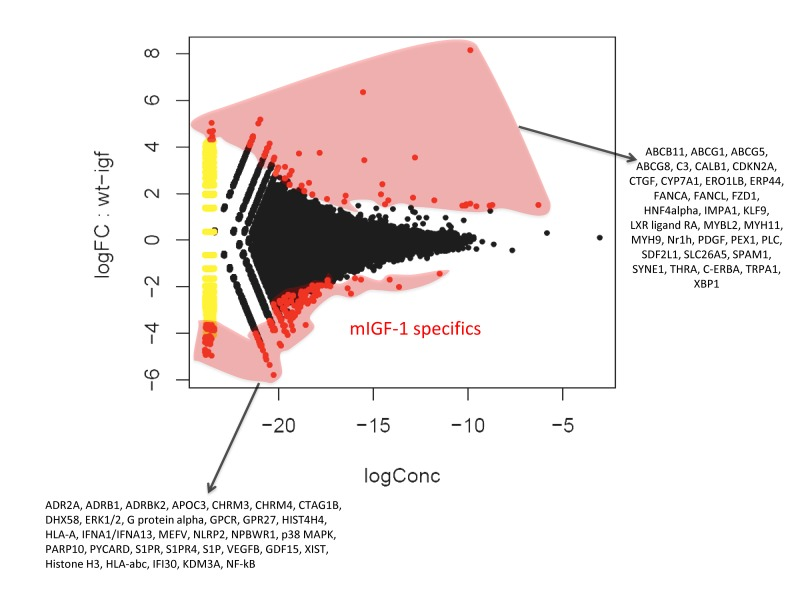
Figure 2. Fold change (FC) plot using common dispersion. The hearts of 2 WT and 2 cardiac-restricted mIGF-1 Tg mice were excised, chromatin was extracted and immunoprecipitated with an anti-SIRT1 antibody. Purified DNA fragments were processed for sequencing with a Solexa Gene Analyzer II platform (Illumina). Upon bioinformatic analyses, 302 TSS were identified as SIRT1 DNA-bound. Red dots represent TSS differentially bound by SIRT1 in the WT or in the mIGF-1 Tg background, while black dots represent TSS bound similarly by SIRT1 in either genotype.
In addition to genome-wide cardiac binding pattern of SIRT1, we sought to determine differences in the transcriptional profiling induced by mIGF-1 transgene in the mouse heart. Previous transcriptomic analyses in the heart of mIGF-1 Tg versus WT mice 24 hours after oxidative injury induced specifically by cardiotoxin injection revealed increased levels of the mitochondrial protein UCP1 (uncoupling protein 1), adiponectin and the antioxidant metallothionein 2 [9]. These three proteins are involved in cardiac protection from oxidative stress, and in isolated cardiomyocytes the upregulation of their levels by mIGF-1 transgene was dependent on SIRT1 activity [5, 9]. Basal transcriptomic analysis on the heart of mIGF-1 Tg versus WT mice in absence of any injury has not been performed and could help to explain why the cardiomyocytes of these mice respond better to specific types of stress (ischemia, angiotensin II, cardiotoxin injection, paraquat injection), with consequent re-generation of the heart tissue [6, 9, 10]. To this purpose, RNA from WT and mIGF-1 Tg hearts was processed for Affymetrix GeneChip analysis. Interestingly, we found that only 22 genes showed a >30% change in mRNA levels (17 upregulated and 5 downregulated, respectively, p<0.05) in the hearts of mIGF-1 Tg mice compared to WT littermates (Fig. 3). Transcript for IGF-1 was found upregulated as expected (Fig. 3, black bar). Most of the upregulated genes in mIGF-1 Tg hearts, 8 out of 17, pertain to the regulation of immune function: lymphocyte antigene 86 (a protein playing an important role in Toll-like receptor 4 pathway), chemokine (C-X-C motif) ligand 14 (a chemoattractant targeting tissue macrophages), C3 (complement component 3, protein whose activation is required for both classical and alternative complement activation pathways, whose promoter is bound by SIRT1, see above), lipopolysaccaride binding protein, CD53 antigen (a tetraspanin protein of the lymphoid-myeloid lineage), CCL21A/CCL21B (potent chemoattractants for lymphocytes) and CCR2 (receptor for monocyte chemoattractant protein-1, MCP-1) (Fig. 3, open bars). The upregulation of these 8 genes was confirmed by qRT-PCR (Fig. 4B) and indicates that cardiac restricted mIGF-1 transgene, in absence of stress, could activate yet uncharacterized immune cell types and mechanisms.
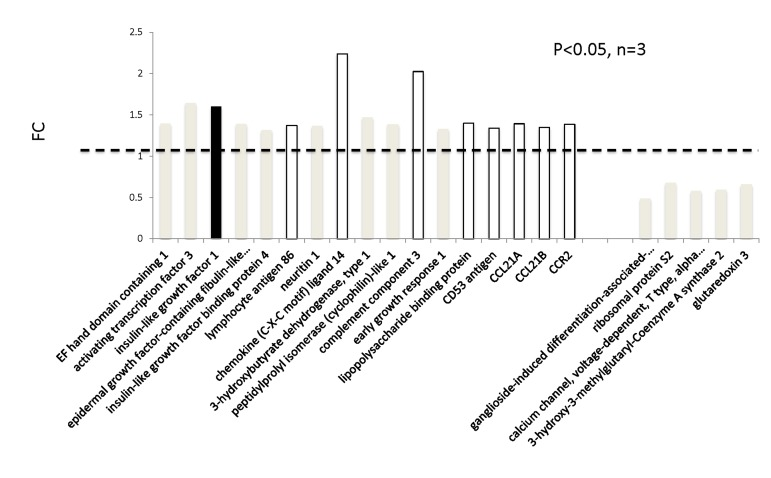
Figure 3. Transcriptomic analyses on the hearts of mIGF-1 Tg versus WT mice. The hearts of 3 WT and 3 cardiac-restricted mIGF-1 Tg mice were excised and RNA was processed for Affymetrix Gene ChIP analyses. Data were analyzed using GeneSpring software. Represented genes are those showing fold changes (FC) starting from 1.3 and a p-value lower than 0.05.
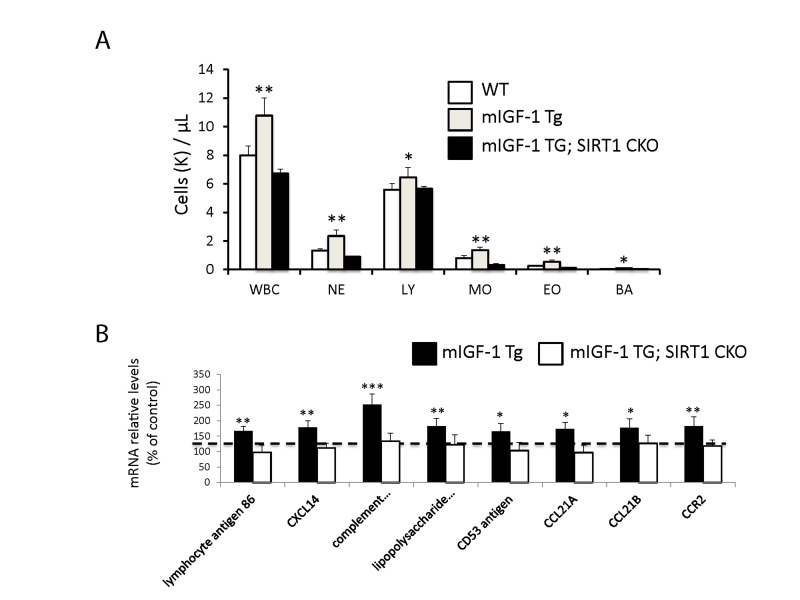
Figure 4. Leukocytosis and cardiac expression of genes involved in the immune response. (A) Blood was extracted from the tail vain of WT, mIGF-1 Tg mice and mIGF-1; SIRT1 CKO mice and was processed for Hemavet analysis to quantify WBC types: neutrophiles (NE), lymphocytes (LY), monocytes (MO), eosinophiles (EO) and basophiles (BA). Results are means ± SE of 5-11 animals for each genotype (*,** p versus WT mice). (B) The expression levels of lymphocyte antigene 86, CXCL14, C3, CD53 antigen, CCL21A, CCL21B and CCR2 mRNAs were examined by Real Time-PCR in the heart of WT, mIGF-1 Tg and mIGF-1 Tg; SIRT1 CKO mice. Results are means ± SE of 5 animals for each genotype (*,**p versus WT mice).
We report thus that at the basal state mIGF-1/SIRT1 genomic and transcriptomic effects in mouse cardiomyocytes are not only related to cardioprotection and anti-oxidant defenses observed under stress conditions [5, 6, 9]. Instead, comparison of both ChIP-Seq data for SIRT1 genome binding and transcriptomic data between WT and mIGF-1 Tg hearts revealed unexpected targets that point to regulatory functions of the mIGF-1/SIRT1 signaling beyond cardiomyocyte specific mechanisms. This led us to perform a more in depth phenotyping and behavioral analysis of these mice, looking closer at three major body compartments involved in stress and aging: the immune system, the arterial blood pressure control system and the behavioral responses dependent on the central nervous system.
Blood leukocytosis and increased arterial blood pressure levels in cardiac restricted mIGF-1 Tg mice
Since ChIP-Seq analysis of SIRT1 genome-wide binding in the mouse heart identified several promoters of genes involved in the immune function, and transcriptomic analysis pointed to an upregulation of 8 key genes implicated in the immune response, we sought to quantify accurately the white blood cell (WBC, leukocytes) number in the peripheral blood of WT and mIGF-1 Tg mice. Blood was collected from WT and mIGF-1 Tg mice through the tail vein and the five different types of leukocytes were measured by hematological profile using the Hemavet blood analyzer. We found that the total count of WBC was significantly increased in the blood of mIGF-1 Tg mice compared to WT mice (Fig. 4A). This total increase in WBC was reflected in a significant increase of all the individual types of leucocytes analyzed (neutrophiles, lymphocytes, monocytes, eosinophiles and basophiles), in terms of thousands of cells per μl (Fig. 4). If values were represented as percentage of cells compared to the total of WBC, mIGF-1 Tg mice displayed 10% less lymphocytes than WT mice, although this difference was not statistically significant (data not shown). To understand if mIGF-1 cardiac transgene induced leukocytosis was dependent on SIRT1 activity, we analyzed the blood of mIGF-1 Tg where SIRT1 activity was specifically ablated in a conditional (αMHC-Cre) and tamoxifen-inducible manner in cardiomyocytes (mIGF-1 Tg; SIRT1 CKO), as we have recently described [6]. This maneuver restored WBC to WT level, indicating that SIRT1 is indispensable for the increase in WBC count observed in cardiac restricted mIGF-1 Tg mice (Fig. 4A). The WBC count of SIRT1 CKO was indistinguishable from WT mice (data not shown). Further, we analyzed by qRT-PCR the cardiac expression of the 8 genes that we found upregulated in Affymetrix analysis in the heart of mIGF-1 Tg mice (lymphocyte antigene 86, CXCL14, C3, CD53 antigen, CCL21A, CCL21B, CCR2) (Fig. 3). A significant upregulation of these genes was confirmed in the heart of mIGF-1 Tg mice compared to WT mice, and it occurred to a similar extent when compared to the one detected by microarrays (Fig. 4B). SIRT1 CKO hearts had similar levels of these transcripts as of WT (data not shown). However, genetic deletion of SIRT1 abrogated the immune gene expression signature observed in mIGF-1 Tg mice (Fig. 4B). These results suggest that SIRT1 is the major transducer whereby mIGF-1 stimulates the expression of immune-related genes in the heart tissue. A raise in total WBC count above the normal range is frequently a sign of an inflammatory response or of a generic stress [25]; moreover, in the elderly leukocytosis has been observed in association to an increased incidence of ischemic cardiovascular events [26]. As stated, mIGF-1 Tg mice are protected from oxidative and hypertrophic stresses and do not display any evident morphological feature of cardiac dysfunction as detected by histology or echocardiography [6, 9]. The latter results, together with the finding that SIRT1 binds promoter of genes involved in cardiovascular function and blood pressure control (ABCG1, C3, CTGF, MYH11, PDGF, TRPA1 in WT hearts; and ADRB1, ERK1/2, G protein alpha, GPCR, p38 MAPK, VEGFB, NF-kB in mIGF-1 Tg hearts) (Fig. 1), prompted us to measure in these animals blood pressure, a direct output of cardiac performance. We non-invasively measured the diastolic and systolic blood pressure (DBP and SBP, respectively) in a cohort of WT, SIRT1 CKO, mIGF-1 Tg and mIGF-1 Tg; SIRT1 CKO using the well-established tail-cuff method. While no difference was observed between WT and SIRT1 CKO mice, these analyses showed a ~20-25% significant increase in both DBP and SBP in mIGF-1 Tg mice compared to WT littermates, in the basal state (Fig. 5). This reproducible increase in diastolic blood pressure (DBP) and systolic blood pressure (SBP) induced by cardiac restricted mIGF-1 transgene could be defined as mild hypertension, as in genetic and dietary murine models of hypertension the values of DBP and/or SBP can reach a 2-fold difference [27]. When blood pressure was analyzed in mIGF-1 Tg; SIRT1 CKO mice, DBP showed a trend of increase compared to WT, although this was not significant and was to a lesser extent when compared to mIGF-1 Tg mice (Fig. 5); SBP on the other hand was found restored to WT level in mIGF-1 Tg; SIRT CKO mice (Fig. 5). These data suggest a mild hypertension in cardiac overexpressing mIGF-1 Tg mice, which could at least in part be rescued by cardiomyocyte-specific inactivation of SIRT1 enzyme.
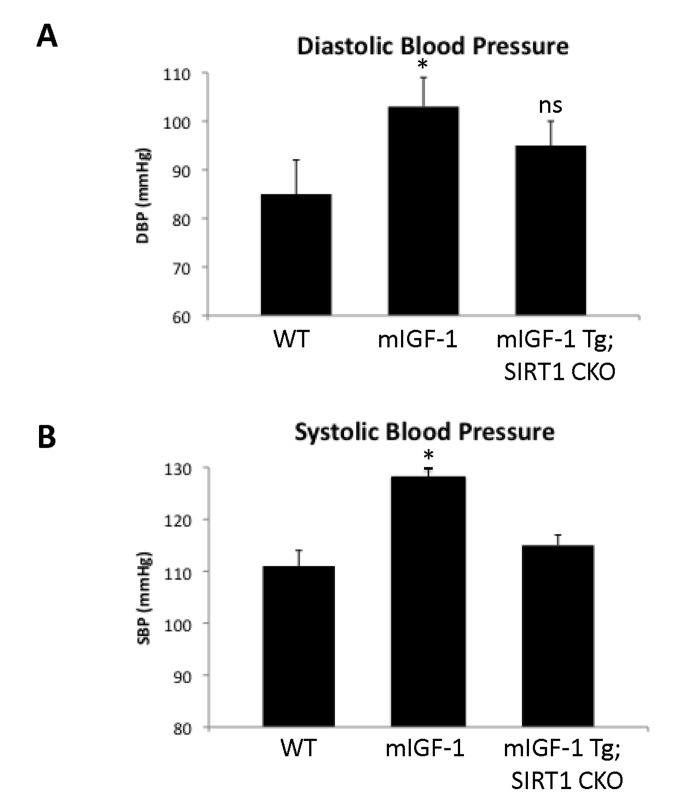
Figure 5. Blood pressure in WT, mIGF-1 Tg and mIGF-1 Tg; SIRT1 CKO mice. Noninvasive blood pressure values in conscious mice were measured using the tail-cuff method. (A) Diastolic blood pressure (DBP); (B) Systolic blood pressure (SBP). Results are means ± SE of 3-8 animals for each genotype (*p versus WT mice). NS= not significant.
Increased response to fear in mIGF-1 Tg mice
It is well established that the effect of stress on the immune system and arterial blood pressure is reciprocally linked to individual and social behavioral responses (such as fear, anxiety) in animal models and human beings [28-30]. To determine if cardiomyocyte specific mIGF-1 transgene could impact on behavioral stress responses, and to understand the role of downstream SIRT1 in this process, we ran in our mice open field and fear conditioning (FC) tests, to determine anxiety levels and responsiveness to an aversive stimulus, respectively. The open field test is commonly used in mice as a qualitative and quantitative measure of locomotor activity and willingness to explore. When WT were compared to mIGF-1 Tg mice, there was no statistical difference in the emotionality/anxiety index if the animals were observed either at the end of the test (30 min, Fig. 6A) or at 5 minute intervals (Fig. 6B). Therefore mild elevated arterial blood pressure in mIGF-1 Tg mice is not overtly linked to anxious behavior. Next, we performed FC test. FC provides a measure of memory of the association between an aversive stimulus such as a mild foot shock and an environmental input. The environmental input consists in a discrete stimulus, i.e. a tone or the test chamber (context). In the FC test, freezing behavior, which is a characteristic fear response in mice (total lack of movement), provides a read-out of memory. Animals that show good memory freeze upon re-presentation of the context (contextual fear conditioning) or the tone (cued fear conditioning). Aging has been reported to decrease memory to an aversive stimulus (decreased freezing) in context- and cue-dependent fear response [31-34]. FC test on WT and mIGF-1 Tg mice showed that mice harboring mIGF-1 transgene in the heart had a better memory of the aversive stimulus, for both cue and context fear response, in that they spent a significant augmented time in a frozen position compared to WT littermates with both environmental inputs (Fig. 6C and D). We then performed FC test in mIGF-1 Tg mice devoid of SIRT1 in cardiomyocytes (mIGF-1 Tg; SIRT1 CKO): these mice surprisingly had a decreased cue-dependent fear response compared to WT, whereas the context-dependent fear response was not significantly altered (Fig. 6C and D). In conclusion, the overexpression of mIGF-1 transgene in mouse heart impinges on the behavior of the whole animals and ameliorates their memory of an aversive stimulus, assessed as freezing in response to fear, and this occurs in a SIRT1 dependent manner.
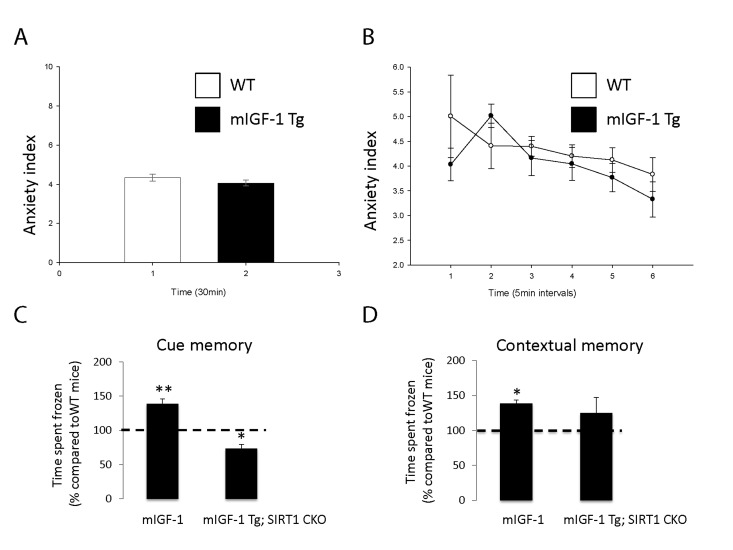
Figure 6. Open field test and fear conditioning test in WT, mIGF-1 Tg and mIGF-1 Tg; SIRT1 CKO mice. (A, B) WT or mIGF-1 Tg mice were subjected to open field test. (C, D) WT, mIGF-1 Tg and mIGF-1 Tg; SIRT1 CKO mice underwent cue memory or conditional memory conditioning tests. Results are means ± SE of 3-8 animals for each genotype (*,**p versus WT mice).