Topology parameters in PPI network from vital targets
Further, all mapped intersection genes were imported to Cytoscape, and then the topological parameters of calycosin-anti-meningitis were determined and reported by using NetworkAnalyzer assays, as shown in candidate genes for data visualization (Figure 2). Following with the assays of Network Statistics and Simple Parameters, the average degree of freedom calculated in the target was 12.837, and the maximum degree of freedom was 49. Therefore, the range of vital target screening conditions was designed as from 26 to 49, and then 7 vital targets were obtained, showing as EGFR, TNF, EGF, ATM, ESR1, CASP8, and NGF (Figure 3).
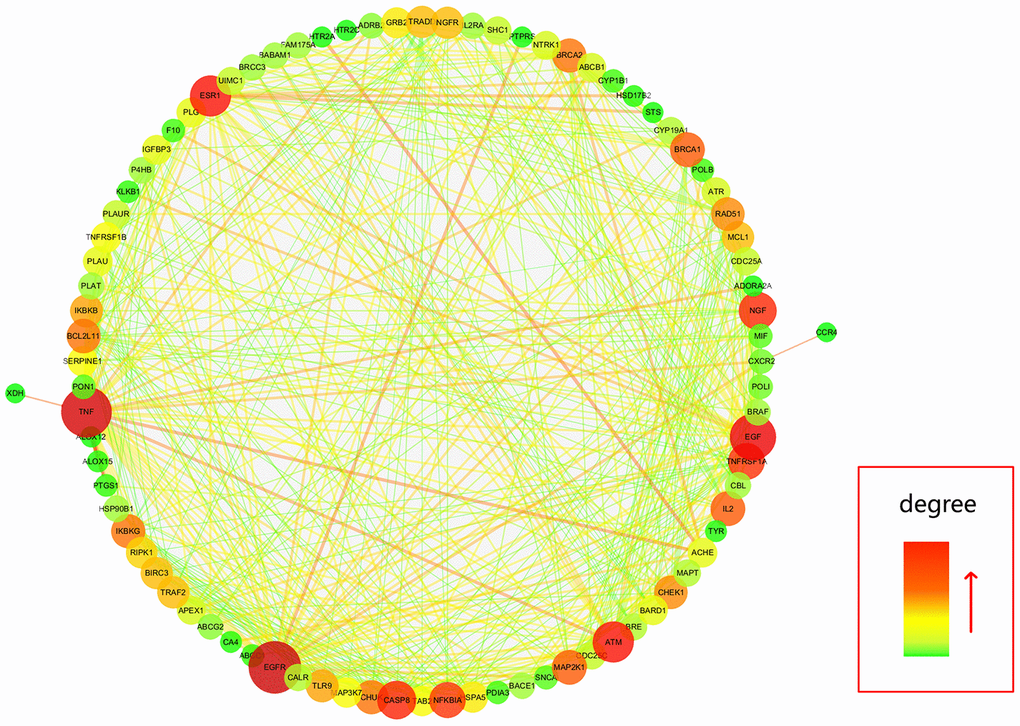
Figure 2. All candidate targets of calycosin-anti-meningitis were collected for construction of a PPI network.
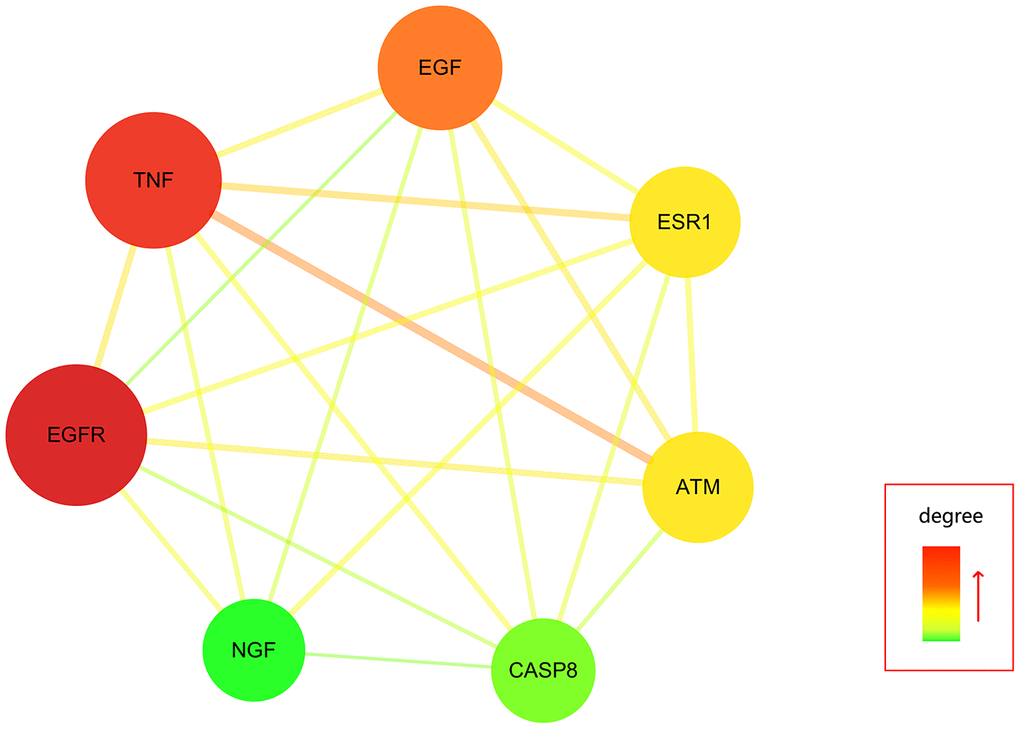
Figure 3. All vital targets of calycosin-anti-meningitis was screened and identified.
Findings of biological functions and pathway enrichment of vital targets
All top KEGG signaling pathway of the vital target were revealed and presented through the DAVID database, and then the important target-related pathway interaction network was showed in Figure 4. The primary signaling pathway of vital targets in calycosin-anti-meningitis were involved in effective regulation of Apoptosis, MAPK signaling pathway, Hepatitis C, FoxO signaling pathway, Proteoglycans in cancer, Rap1 signaling pathway, Ras signaling pathway, PI3K-Akt signaling pathway, Bladder cancer, Pathways in cancer, Endometrial cancer, Legionellosis, NOD-like receptor signaling pathway, Non-small cell lung cancer, Pancreatic cancer, Glioma, p53 signaling pathway, RIG-I-like receptor signaling pathway, Melanoma, NF-kappa B signaling pathway, ErbB signaling pathway, Prostate cancer, Gap junction, HIF-1 signaling pathway, Estrogen signaling pathway, Choline metabolism in cancer, Chagas disease (American trypanosomiasis), Toll-like receptor signaling pathway, TNF signaling pathway, Toxoplasmosis (more details shown in Supplementary Figure 1). Furthermore, the biological functions of vital targets were identified through the DAVID database, as revealed in positive regulation of nitric oxide biosynthetic process, activation of MAPKK activity, positive regulation of MAP kinase activity, activation of cysteine-type endopeptidase activity involved in apoptotic process, phosphatidylinositol-mediated signaling, regulation of apoptotic process, death-inducing signaling complex assembly, MAPK cascade, positive regulation of catenin import into nucleus, positive regulation of apoptotic process, signal transduction, mammary gland alveolus development, positive regulation of phosphorylation, regulation of cell motility, regulation of tumor necrosis factor-mediated signaling pathway, cellular response to estradiol stimulus, negative regulation of epidermal growth factor receptor signaling pathway, positive regulation of transcription, DNA-templated, extrinsic apoptotic signaling pathway via death domain receptors, ERBB2 signaling pathway, negative regulation of I-kappaB kinase/NF-kappaB signaling, extrinsic apoptotic signaling pathway, cellular response to amino acid stimulus, intrinsic apoptotic signaling pathway in response to DNA damage, neuron projection morphogenesis, positive regulation of fibroblast proliferation, epidermal growth factor receptor signaling pathway, cellular response to organic cyclic compound, positive regulation of smooth muscle cell proliferation, cellular response to mechanical stimulus, regulation of phosphatidylinositol 3-kinase signaling, positive regulation of protein kinase B signaling, response to estradiol, phosphatidylinositol phosphorylation, transmembrane receptor protein tyrosine kinase signaling pathway, positive regulation of sequence-specific DNA binding transcription factor activity, activation of MAPK activity, positive regulation of transcription from RNA polymerase II promoter, positive regulation of protein phosphorylation, negative regulation of gene expression, peptidyl-tyrosine phosphorylation, DNA replication, positive regulation of I-kappaB kinase/NF-kappaB signaling, protein autophosphorylation, positive regulation of ERK1 and ERK2 cascade, positive regulation of gene expression, cell surface receptor signaling pathway (more details shown Supplementary Table 1). More markedly, the top 20 functional processes and molecular pathways of calycosin-anti-meningitis were optimized and highlighted (Figure 5). Collectively, we concluded that the bioinformatics flow-scheme by using network pharmacology was demonstrated in Figure 6.
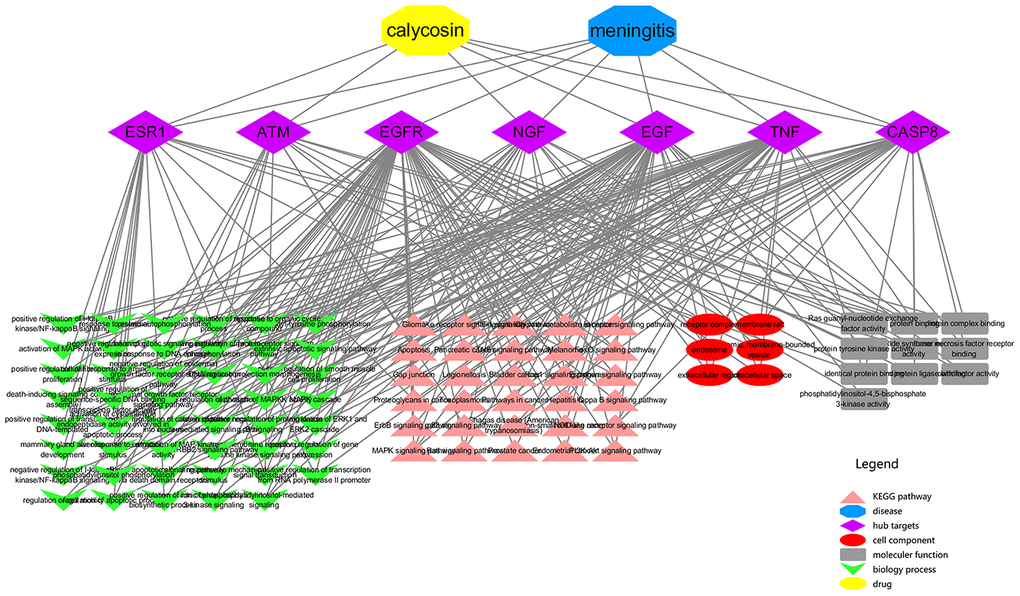
Figure 4. An integrated network from vital targets was plotted and revealed the intersection association of target-disease-function-pathway in calycosin-anti-meningitis.
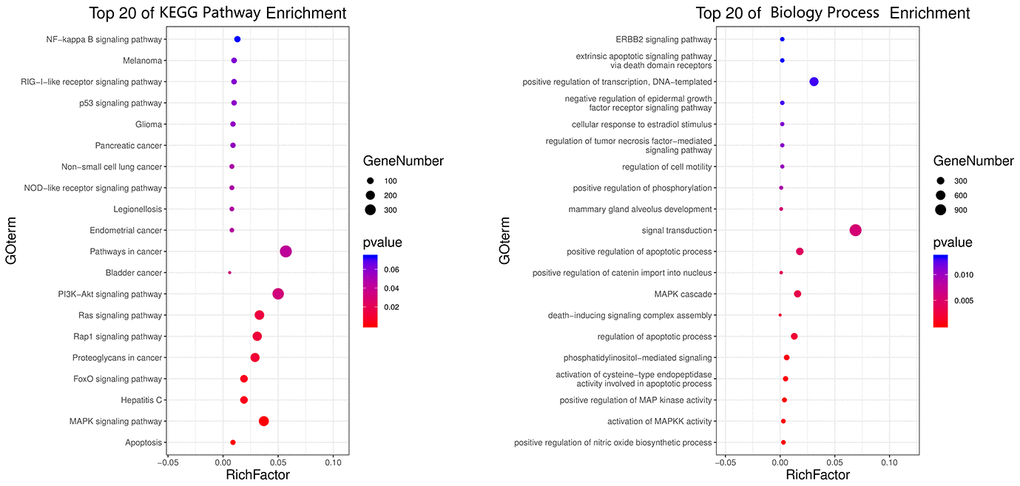
Figure 5. All top 20 biological processes and molecular pathways of calycosin-anti- meningitis from enrichment analyses were revealed and visualized.
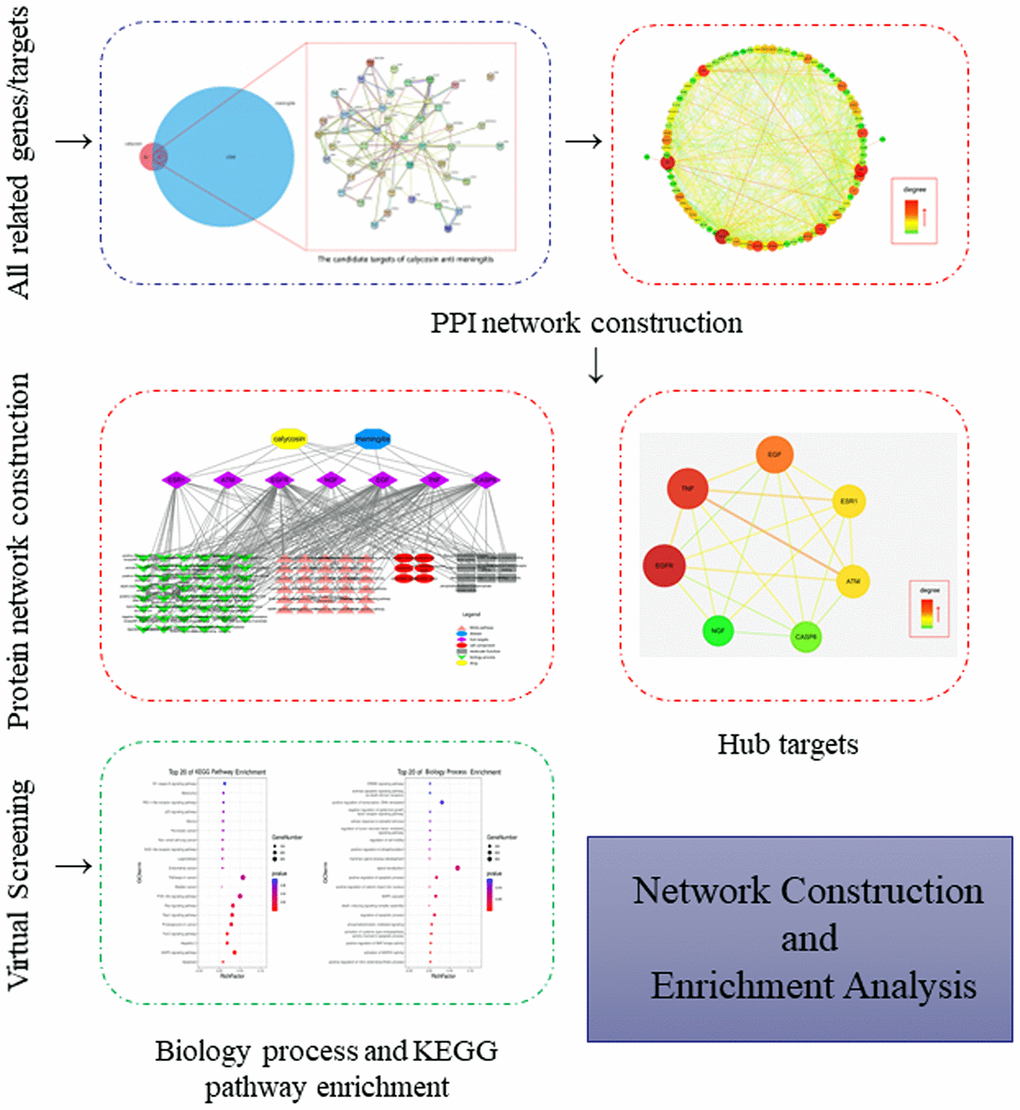
Figure 6. A flow-chart using a network pharmacology approach aimed to reveal and characterize the biological targets, functions and molecular pathways in calycosin-anti-meningitis.
Molecular docking findings
In the EGFR protein, the RMSD of the original ligand was 3.889 Å. The hydrogen bonding between the pro-ligand 8BS and the 5UGC protein acted on the amino acid residue with MET-793 (1.9Å), and the free binding energy with protein was -8.06 kcal/mol. Calycosin formed hydrogen bond with amino acid residue LYS-745 (2.4Å), and the free binding energy with protein is -8.9 kcal/mol (Figure 7A). In the TNF protein, the RMSD of the original ligand was 2.375 Å. The hydrogen bonding between the pro-ligand A7M and the 6OOY protein acted on the amino acid residue with SER-60 (2.9Å), TYS-151(1.8Å), and the free binding energy with protein was -4.87 kcal/mol. Calycosin formed hydrogen bond with amino acid residue TYS-151 (2.2Å), and the free binding energy with protein is -6.7 kcal/mol (Figure 7B).
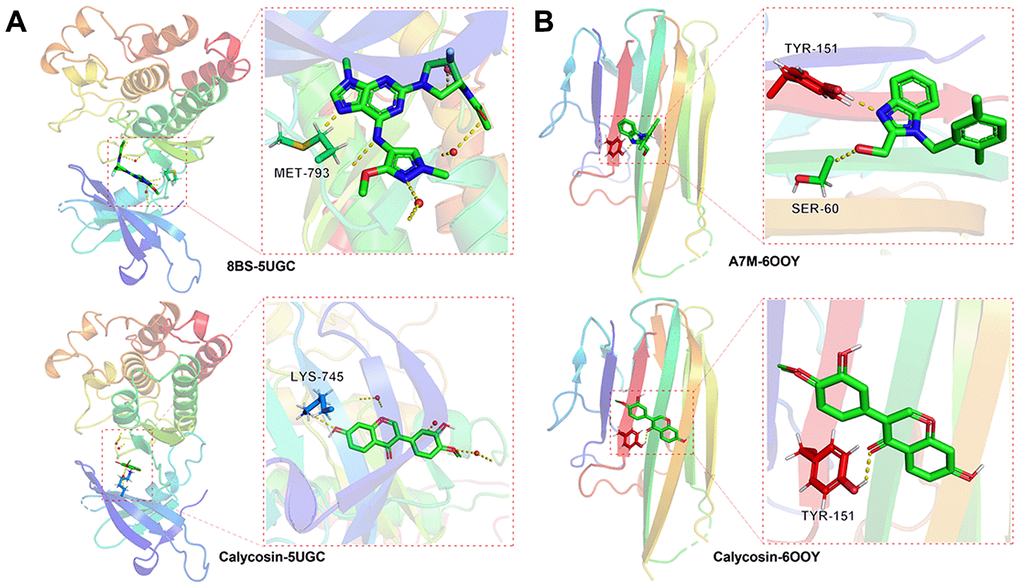
Figure 7. Molecular docking data indicated that the binding capacity of calycosin with meningitis was significant in the vital targets of EGFR, 8BS-5UGC (A), TNF, A7M-6OOY (B).