Abstract
The developmental origins of health and disease concept links adult diseases with early-life exposure to inappropriate environmental conditions. Intrauterine and postnatal malnutrition may lead to an increased incidence of type 2 diabetes, obesity, and cardiovascular diseases. Maternal malnutrition (MM) has also been associated with prostate carcinogenesis. However, the molecular mechanisms associated with this condition remain poorly understood. Using a proteomic analysis, we demonstrated that MM changed the levels of proteins associated with growth factors, estrogen signaling, detoxification, and energy metabolism in the prostate of both young and old rats. These animals also showed increased levels of molecular markers of endoplasmic reticulum function and histones. We further performed an in silico analysis that identified commonly deregulated proteins in the ventral prostate of old rats submitted to MM with a mouse model and patients with prostate cancer. In conclusion, our results demonstrated that estrogenic signaling pathways, endoplasmic reticulum functions, energy metabolism, and molecular sensors of protein folding and Ca2+ homeostasis, besides histone, and RAS-GTPase family appear to be involved in this process. Knowledge of these factors may raise discussions regarding the role of maternal dietary intervention as a public policy for the lifelong prevention of chronic diseases.
Introduction
Advances in public health and preventive medicine have resulted in an unprecedented and welcomed number of individuals reaching old age. Despite longevity, there has been observed an increase in older populations affected by chronic diseases that demand specialized and expensive elderly care services. The identification and widespread public awareness of unhealthy modifiable risk factors such as western diet consumption and obesity, sedentary lifestyle, smoking, stress, and insufficient sleep, followed by the adoption of a healthy lifestyle is a feasible, safe, and effective low-cost public policy program to improve the quality of life with aging [1, 2]. Intrauterine and early postnatal life experiences may permanently modulate health trajectories across the lifespan [3–5]. The hypothesis that the intrauterine period of development may modulate offspring postnatal health was initially proposed by David Barker in "Fetal Origin of Adult Diseases" (FOAD), almost 30 years ago [6]. Subsequently, FOAD evolved to consolidate the "Developmental Origins of Health and Disease" (DOHaD) concept by including both pre-conception and early postnatal life as a new window of susceptibility. Recently, epigenetics has become one of the most relevant molecular mechanisms associated with the transgenerational inheritance involved with DOHaD [7].
Despite the difficulty in confirming the impact of maternal and early life adversity on human health, some tragic events in human history were crucial in supporting the DOHaD concept. For example, the "Dutch Hunger Winter" was a period of severe famine in the western part of the Netherlands at the end of World War II and has provided an opportunity to explore the effects of intrauterine malnutrition on subsequent adult health [8]. First published in 1976, the Dutch Hunger Winter Cohort has been explored to confirm the developmental origin of non-communicable chronic diseases, e.g., cardiovascular disease, obesity, type 2 diabetes, schizophrenia, and infertility in the progeny exposed to famine [9–11]. Importantly, epidemiological and experimental studies have also supported the DOHaD concept as a mechanistic framework related to early life carcinogenesis (such as breast and prostate cancer) [12–17]. Among the malignancies affecting men, prostate cancer (PCa) is the second most diagnosed cancer worldwide. In 2018, the Global Cancer Statistics (GLOBOCAN) estimated almost 1.3 million new cases of PCa globally, leading to 359,000 deaths [18]. Although multifactorial etiology, genetic background, ethnicity, and aging are consistently established risk factors for PCa. However, evidence supporting the early origin of PCa is growing. William Gardner (1995) proposed, almost 30 years ago, the "Prenatal origin of PCa" hypothesis [19]. After that, some epidemiological studies have reinforced Gardner's hypothesis on the early life origins of PCa, as diagnosed in older men [14, 20, 21]. These authors proposed that exposure to certain environmental conditions during pregnancy, such as malnutrition or chemical endocrine disruptors, may alter maternal steroid hormone profiles, thereby modifying the offspring's PCa risk throughout life. This effect was consistently observed in African American men, who are at high risk for PCa, and whose mothers have higher levels of estrogen during pregnancy compared to Caucasian women [20].
In one of the few opportunities to explore how exposure to adverse conditions during windows of vulnerability interferes with human PCa, Keinan-Boker et al. [13] demonstrated that Jewish men exposed during early life to famine and stress during the Holocaust were at a higher risk for several types of cancer (including PCa) later in life. Similarly, women severely exposed to famine during the Dutch hunger winter were at increased risk for breast cancer development [22]. Interestingly, a higher risk for breast cancer was observed for women who were exposed to famine between the ages of 2 and 9 years. Dirx et al. [23] analyzing data from the Netherlands Cohort Study, showed a slight increase in PCa risk among men exposed to famine during adolescence (a critical window of vulnerability for reproductive organs) compared with those men living in northern and southern parts of the Netherlands, who had almost no exposure to famine. These data reinforce the need to explore, in-depth, the potential of early life malnutrition as an environmental risk factor for prostate carcinogenesis across the lifespan.
Emerging experimental studies have been designed to explore the potential of early life exposure to environmental risk factors on prostate carcinogenesis. The intrauterine or neonatal exposure to endocrine disruptors, such as phthalate or bisphenol A, has been associated with the deregulation of critical molecular pathways involved in prostate carcinogenesis in rat offspring [17, 24, 25]. Regarding maternal malnutrition (MM), Santos et al. [16] demonstrated that offspring born from dams fed with a low protein diet (LPD) during gestation and lactation were at high risk of developing prostatic disorders with aging, including carcinoma in situ in the ventral prostate (VP) lobe. Overall, it has been proposed that early life exposure to endocrine disruptors or malnutrition may show early deregulation and persistent cellular response to estrogen signaling pathways, probably involving changes in epigenetic markers, such as DNA methylation and the expression of microRNAs, leading to an increased incidence of prostatic disorders with aging [17, 25–27].
Although MM can be identified as a potentially modifiable environmental risk factor for offspring prostate carcinogenesis, there is a lack of information regarding the molecular mechanisms involved in this process. We previously demonstrated, for the first time, that maternal exposure to a low protein diet promotes prostate carcinogenesis in older rat offspring [16]. In the current investigation, we used mass spectrometry to identify, in young and older offspring, changes in the proteomic profile potentially involved in the early life origins of prostate carcinogenesis observed with aging.
Results
Maternal LPD reduced weight gain and imbalance of steroid hormones in male offspring
Offspring body weight was lower in the LPD animals on both postnatal days (PND) 21 and 540 compared to the respective control (CTR) groups (Figure 1A, 1B). The serum steroid hormones estrogen (17β-estradiol) and testosterone (17β-hydroxy-4-androstene-3-one) increased in the GLLP group on PND 21 compared to the CTR group (Figure 1C). However, on PND 540, while estrogen was higher in the GLLP group, testosterone levels decreased compared to the CTR group, leading to an increased estrogen/testosterone ratio (Figure 1D).
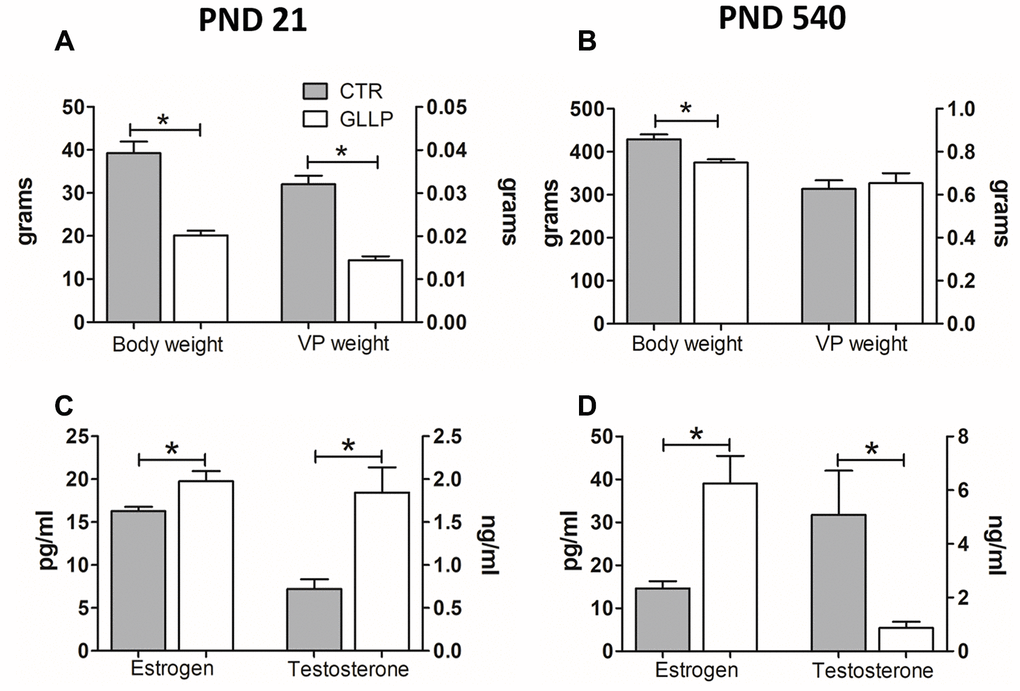
Figure 1. Body weight (A, B) and hormonal levels (C, D) of male offspring on PND 21 and 540. All data are expressed as mean±SD. Asterisks (*) represent statistical differences between experimental groups with p<0.05. CTR = control; GLLP = gestational and lactational low protein; PND = postnatal day; VP = ventral prostate.
Early and late effects of maternal LPD on offspring VP
On PND 21, the morphological analyses demonstrated an impairment of prostate growth in the GLLP group, characterized by a smaller prostatic secretory structure, reduced luminal compartment, and increased epithelial and stromal compartments, compared to the CTR group (Figure 2A, 2B). On PND 540, while we did not identify carcinoma in the CTR group (Figure 2C), the histopathological analysis confirmed the presence of carcinoma in situ in the animals from the GLLP group selected for a mass spectrometry analysis (Figure 2D).
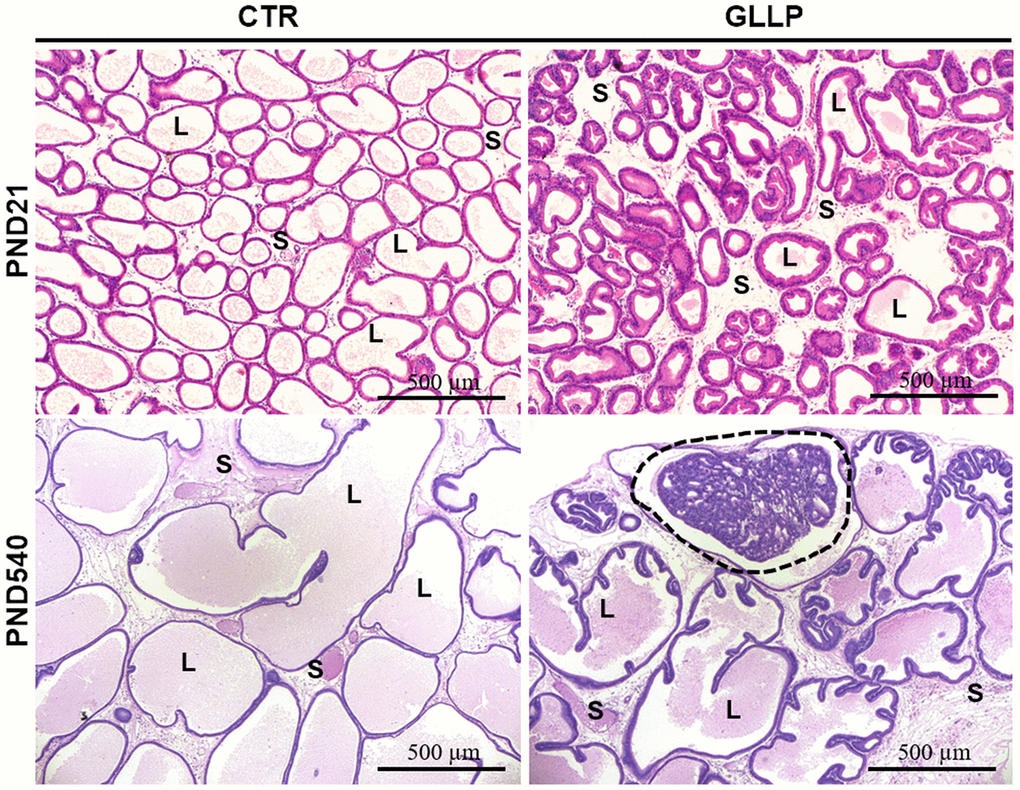
Figure 2. Representative histological sections of the VP lobes from the CTR and GLLP groups on PND 21 and 540, stained with hematoxylin-eosin (HE). Glandular growth in the GLLP group on PND 21 was impaired compared to the CTR. At PND 540, the carcinoma in situ was highlighted by the dashed circle. S = Stroma, L = Lumen, E = Epithelium, Scale bar: 500 μm.
Maternal LPD changed the proteomic profile in the prostate offspring at both ages
Figure 3 shows a total of 256 proteins identified in the VP by MS/MS approach on PND 21. Of these, 158 proteins were significantly differentially expressed in the GLLP group compared to the CTR group, including 138 and 20 proteins that were up- and downregulated, respectively. On PND 540, 366 proteins were significantly differentially expressed in the GLLP group compared to the CTR group, including 135 and 141 proteins that were up- and downregulated, respectively. The complete list of proteins is described in Supplementary File 1.
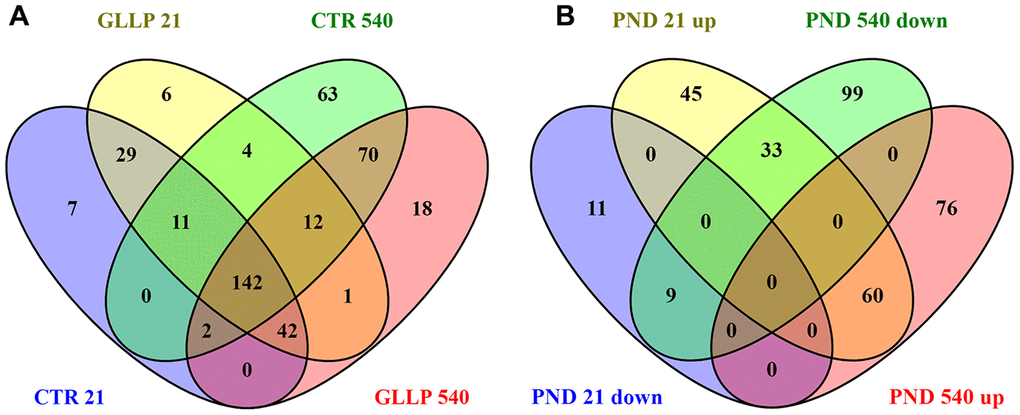
Figure 3. Venn diagram. (A) Shared proteins between CTR and GLLP groups in PND 21 and 540. (B) Shared proteins differentially expressed between the CTR and GLLP groups on PND 21 and 540. CTR = Control; GLLP = Gestational and lactational low protein; PND = Postnatal day; up = upregulated proteins; down = downregulated proteins.
Functional enrichment identified molecular pathways altered by maternal LPD in the offspring prostate
Functional enrichment was performed for the set of downregulated and upregulated proteins separately in both ages. The red and blue bars in Figure 4A demonstrate enriched terms for up and downregulated proteins on PND 21, respectively. Upregulated proteins enriched terms related to protein processing in the endoplasmic reticulum, antigen processing and presentation, metabolism of xenobiotics by cytochrome P450, endocytosis, estrogen signaling pathway, longevity regulating pathway - multiple species, apoptosis signaling pathway, chemical carcinogenesis, spliceosome, glutathione metabolism, arachidonic acid metabolism, drug metabolism - cytochrome P450, and MAPK (mitogen-activated protein kinase) signaling pathways. Downregulated proteins enriched terms related to cell cycle, Hippo signaling pathway, FGF (fibroblast growth factor) signaling pathway, EGF (epidermal growth factor) receptor signaling pathway, cytoskeletal regulation by Rho GTPase, apoptosis, cadherin signaling pathway, gap junction, phosphatidylinositol 3' -kinase (PI3K)-AKT signaling pathway, regulation of actin cytoskeleton, tight junction, focal adhesion, RAP1 (Ras-proximate-1) signaling pathway, adherents junction, and p53 signaling pathway. Using a circus plot analysis, we identified the set of deregulated proteins (up and down) associated with each enriched term on PND 21 (Figure 4B). On PND 540, the upregulated proteins enriched terms related to the metabolism of xenobiotics by cytochrome P450, chemical carcinogenesis, glutathione metabolism, drug metabolism - cytochrome P450, platinum drug resistance, and protein processing in the endoplasmic reticulum. Downregulated protein-enriched terms related to cytoskeletal regulation by Rho-GTPase, phagosome, gap junction, glycolysis/gluconeogenesis, biosynthesis of amino acids, carbon metabolism, fructose galactose metabolism, regulation of actin cytoskeleton, glycolysis, and cadherin signaling pathway (Figure 5A). Using a circus plot analysis, we identified the set of deregulated proteins (up and down) associated with each enriched term on PND 540 (Figure 5B). The list of proteins that enriched each molecular term is described in Supplementary File 2.
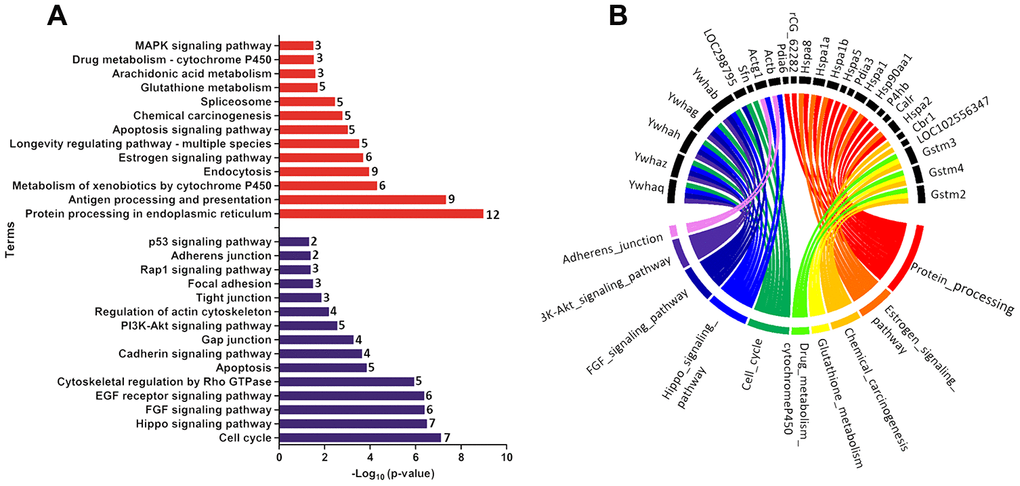
Figure 4. (A) Ontological enrichment of upregulated (red) and downregulated (blue) proteins on PND 21 by KOBAs 3.0. All data were expressed as -Log10 (p-value). (B) Circus plot graphic identifying the top 10 enriched terms and the DEP associated with each term. The numbers in front of the bars mean the number of proteins that enriched each term.
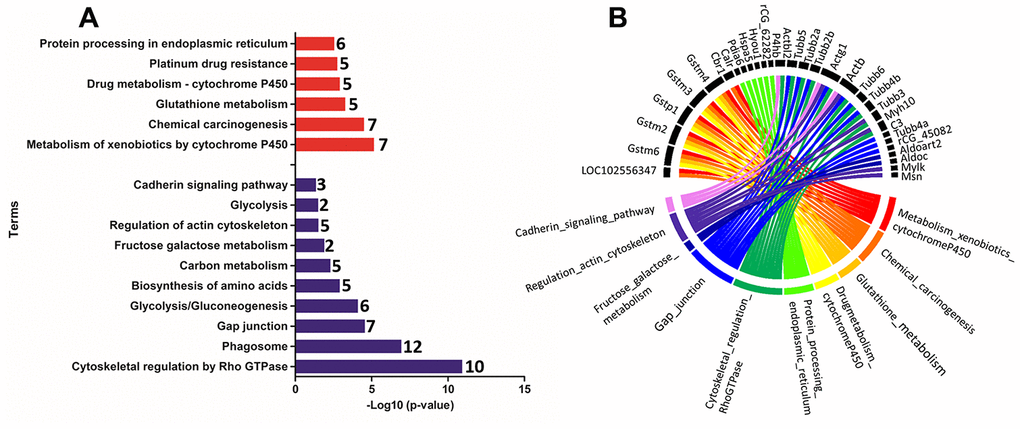
Figure 5. (A) Ontological enrichment of upregulated (red) and downregulated (blue) proteins on PND 540 using the KOBAs 3.0 tool. All data were expressed as -Log10 (p-value). (B) The Circus plot graphic identifying the top 10 enriched terms and the DEP associated with each term.
The enrichment analysis from Ligand Perturbation UP/DOWN database showed that upregulated proteins (in both PND 21 and 540) are associated with hormonal treatment, especially testosterone and estrogen (Figure 6A, 6B). The set of downregulated proteins for both ages also enriched terms related to exposure to testosterone and estrogen (Figure 7A, 7B). Overall, these results highlighted the involvement of a hormonal imbalance on maternal LPD-induced prostate disorders in offspring.
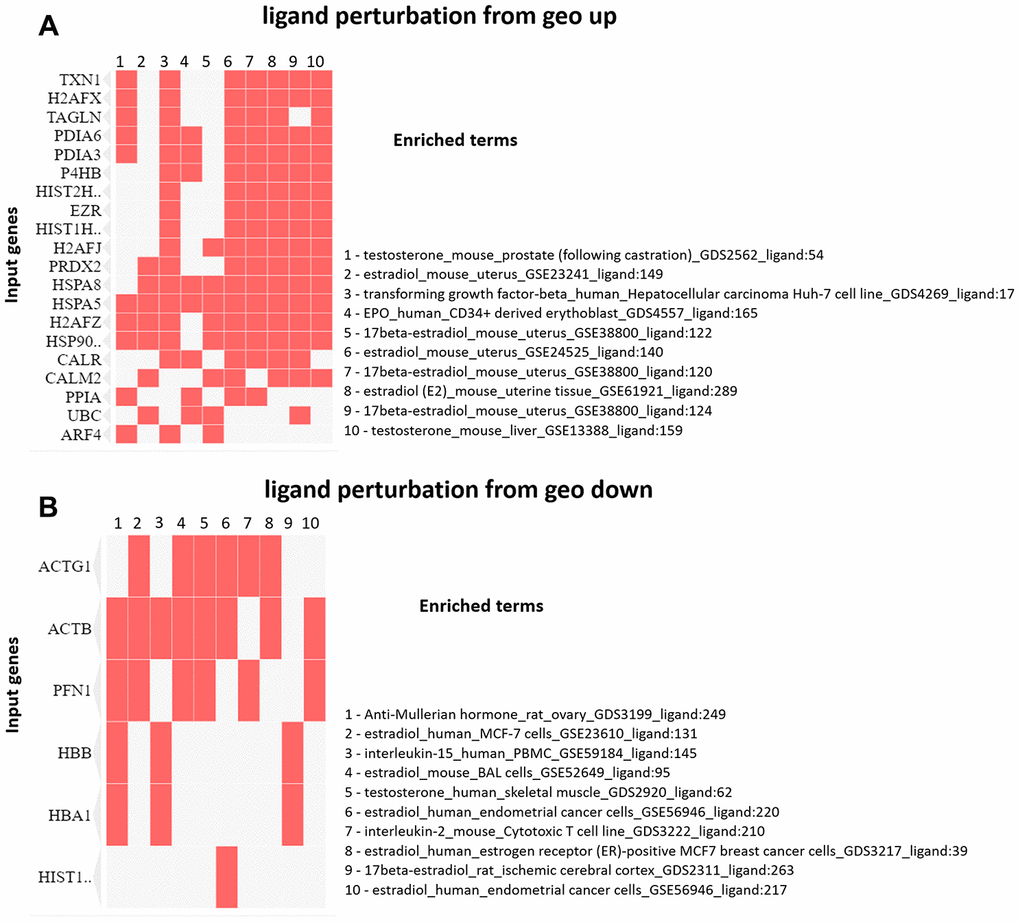
Figure 6. (A) Clustergram generated by Enrichr using upregulated proteins on PND 21. The red cells in the matrix indicate the genes associated with each term. It was demonstrated the top 10 enriched terms with p-value <0.05. (B) Clustergram generated by Enrichr using downregulated proteins on PND 21. The red cells in the matrix indicate the genes associated with each term. It was demonstrated the enriched terms with p-value <0.05 (top 10).
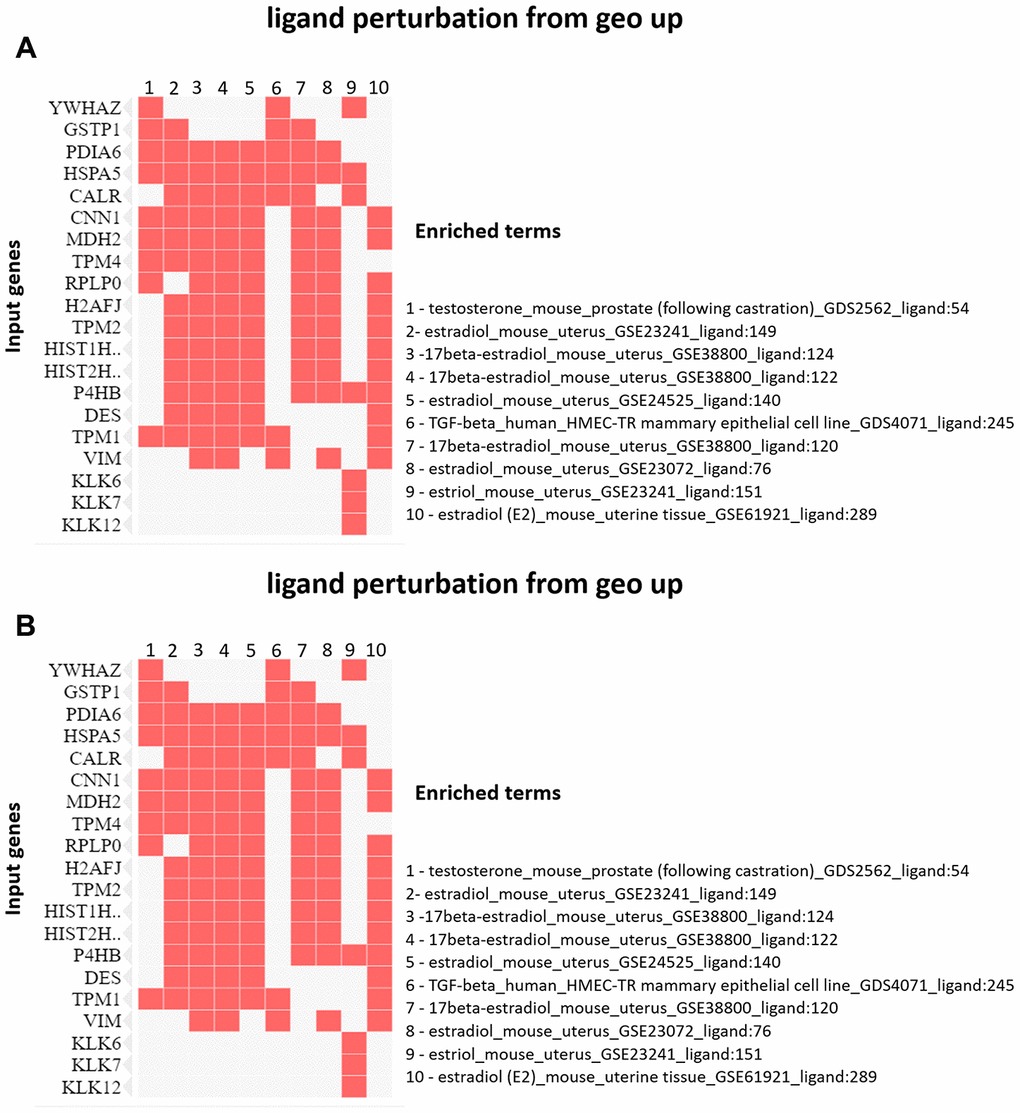
Figure 7. (A) Clustergram generated by Enrichr using upregulated proteins on PND 540. The red cells in the matrix indicate the genes associated with each term. It was demonstrated the enriched terms with p-value <0.05 (top 10). (B) Clustergram generated by Enrichr using downregulated proteins on PND 540. The red cells in the matrix indicate the genes associated with each term. It was demonstrated the enriched terms with p-value <0.05 (top 10).
Protein-Protein Interaction network
Protein-Protein interaction (PPI) network analysis demonstrated several clusters for up and downregulated proteins on PND 21 and 540 (Supplementary Figures 1–4). Based on these results, we identified five principal clusters commonly deregulated in both ages: (1) RAB (Ras-related protein) 1, RAB10, RAB15, RAB1A, RAB35, RAB8A, AND RAB8B; (2) H2AFJ (Histone H2A.J), HIST1H2AA, HIST1H2AH, HIST1H2AK, HIST1H2AN, HIST2H2AC, and HIST3H2A; (3) GSTM2 (glutathione S-transferase Mu 2), GSTM4 and GSTM7; (4) CALR (calreticulin), HSPA5 (heat shock protein family A member 5), P4HB (protein disulfide isomerase-4), PDIA6 (Protein Disulfide Isomerase Family A Member 6); (5) PRDX5 (peroxiredoxin-5) and TXN1 (thioredoxin 1). The cluster identified in commonly downregulated proteins on PND 21 and 540 was composed of HBA1 (hemoglobin Subunit Alpha 1), HBA-A2, HBB, and HBE1 (Figure 8).
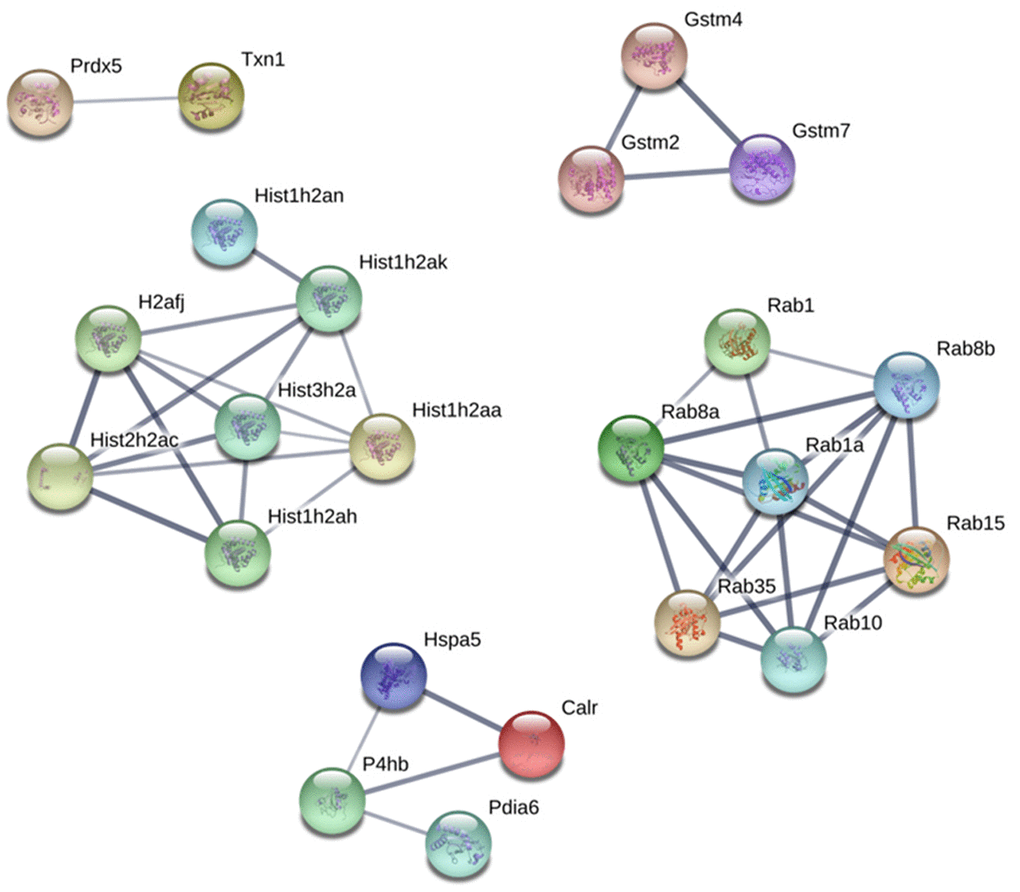
Figure 8. Protein-protein interaction network between commonly upregulated proteins on both PND 21 and 540. Interactions of the identified proteins were mapped by searching the STRING database version 9.0 with a confidence cut-off of 0.7. In the resulting protein association network, proteins are presented as nodes that are connected by lines whose thickness represents the confidence level (0.7-0.9).
In silico analysis confirmed the relationship between differentially expressed proteins (DEP) and PCa in both rodent model and human samples
To give further insights into the role of maternal malnutrition on prostate carcinogenesis, we compared our set of DEP with data from a transgenic PCa mouse model and data from The Cancer Genomic Atlas (TCGA) patients with PCa taken from Gene Expression Profiling Interactive Analysis (GEPIA). In the PB-Cre/PtenloxP/loxP, we identified a set of DEP commonly expressed in our samples, mainly on PND 540 and in the prostatic tumors in all prostatic lobes. Interestingly, the percentage of commonly deregulated targets between our samples and those from the PB-Cre/Ptenl oxP/loxP model increased with aging (from PND 21 to 540) and with the aggressiveness of prostatic lesions (PND 21: PIN 3.6%; medium 5.0%; advanced 6.8% and PND 540: PIN 13.1%; medium 15.9%; advanced 21.3% (Figure 9A). Similar results were obtained when our set of DEP was compared with data from patients with PCa (from 5.9% on PND 21 to 12.0% on PND 540) (Figure 9B). We also identified six proteins (CALR, HIST2H2AC, HSPA5, P4HB, and PDIA6) in the HPA database that showed increased immunostaining in PCa tumor tissue, while low or not detected in normal prostate tissue (Figure 9C). These results highlight the involvement of maternal malnutrition in the deregulation of proteins involved in prostate tumors.
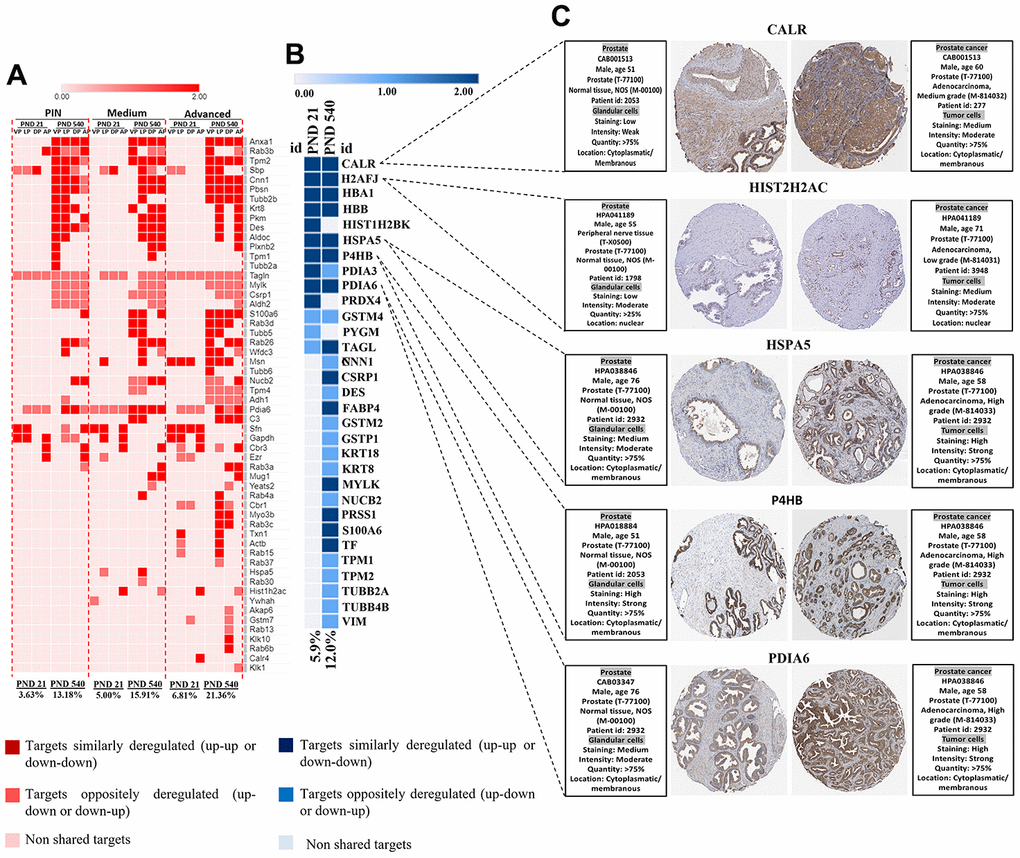
Figure 9. (A) Heatmap showing the commonly differentially targets from our set of DEP and RNA-seq data from ventral, dorsal, lateral, and anterior prostate lobes in the mice model of PCa (PB-Cre/PtenloxP/loxP). The percentage of commonly deregulated targets increases as the prostatic disorders worsen (PIN to Medium to Advanced PCa). (B) The commonly deregulated targets between our DEP and those extracted from RNA-seq data by GEPIA. The percentage of commonly deregulated targets increases in the prostate of older offspring. (C) Immunostaining of normal and prostate tumor samples for five commonly upregulated targets in our samples and GEPIA database (http://gepia.cancerpku.cn/) using immunohistochemical data available at the Human Protein Atlas database (https://proteinatlas.org/). PND: Postnatal day; VP: Ventral prostate; LP: Lateral prostate; DP: Dorsal prostate; AP: Anterior prostate PIN: Prostate intraepithelial neoplasia; PCa: Prostate cancer.
Experimental validation of CALR as upregulated protein in offspring exposed to maternal malnutrition
Based on the proteomic data (Supplementary File 1) and in silico analysis, we employed immunohistochemical and RT-qPCR analyses to validate the CALR as an upregulated target in the offspring VP born to dams fed with LPD in both PND 21 and 540. Immunostaining for CALR was more evident in the GLLP group on both PND 21 (Figure 10C and 10D) and PND 540 (Figure 10G, 10H) compared to the CTR group (Figure 10A, 10B, 10E, 10F) mainly in areas of carcinoma in situ. RT-qPCR confirmed the upregulation of CALR gene expression in the GLLP group at both ages (Figure 10I, 10J).
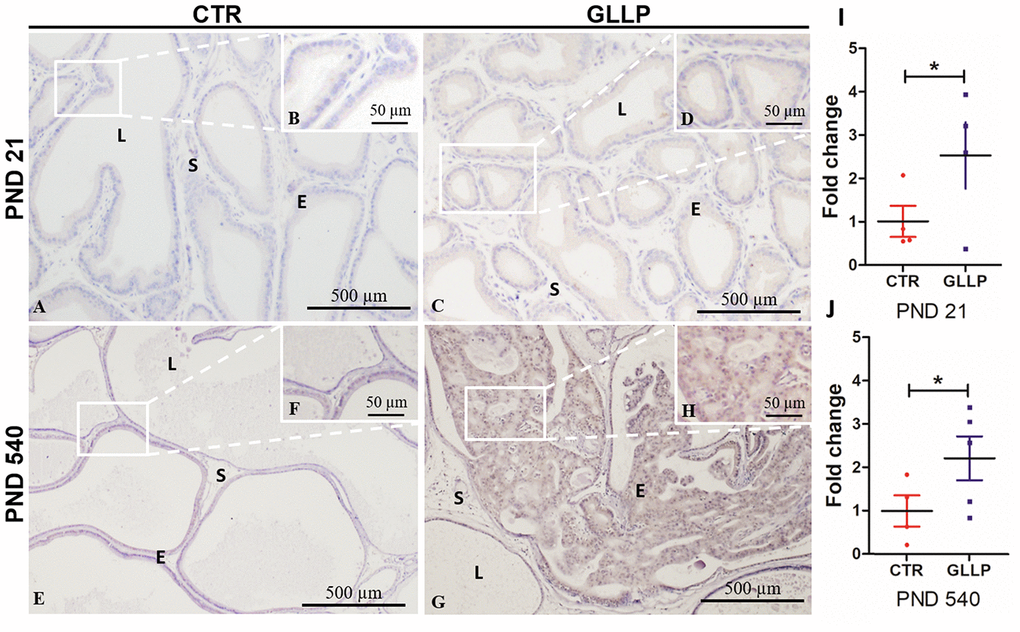
Figure 10. Representative immunohistochemistry reaction for Calreticulin (CALR) in the VP lobes from the CTR and GLLP groups on PND 21 (A–D) and 540 (E–H). RT-qPCR reaction for CALR in the VP lobes from CTR and GLLP groups on PND 21 (I) and 540 (J). CTR = control; GLLP = gestational and lactational low protein; PND = postnatal day. Data are expressed as fold change {plus minus} SD. Asterisks (*) means the statistical difference between experimental groups with p < .05. Scale bar: 500 μm, and detail 50 μm.
Discussion
Although maternal exposure to adverse conditions has been identified as an essential window for the development of non-communicable diseases in the progeny, there is a growing body of evidence highlighting malnutrition during early life as a key environmental risk factor for the developmental origin of some types of diseases such as some types of cancer, including breast and PCa in offspring [12–16, 18, 28–30]. However, little data is supporting the molecular pathways associated with early life carcinogenesis and understanding this aspect may be crucial to identifying and perhaps modulating molecular pathways involved in the developmental origin of diseases, especially in those more vulnerable populations, who have limited access to more expensive food components, such as proteins [31].
Consistent with our previous results [16, 32], the set of deregulated proteins identified in a mass spectrometry analysis was associated with the molecular mechanism classically recognized as a potent regulator of development, maintenance of tissue homeostasis, and disease. FGFs and EGFs act to regulate glandular morphogenesis, cell proliferation, and differentiation and have secretory functions not only during development but also during neoplastic transformation and tumor progression [33–38]. The Hippo signaling pathway also plays a crucial role in the control of organ size, branching morphogenesis, and tissue homeostasis by regulating cellular mechanisms such as cellular polarity, cell-cell contact, and cytoskeleton organization [39, 40]. The enrichment of molecular pathways related to the endoplasmic reticulum function, such as the metabolism of xenobiotic, chemical carcinogenesis, endocytosis, in addition to cell adhesion, longevity, and apoptosis also highlights the involvement of these cellular and molecular mechanisms in prostatic carcinogenesis [41–45]. The maintenance of these processes is crucial for glandular homeostasis. As a consequence, the breakdown of these molecular mechanisms may also interfere with cell-cell adhesion and metabolism, the maintenance of barriers between the blood and the epithelial and stromal compartments, in addition to interfering within the dynamics of cellular differentiation, proliferation, and migration, all mechanisms involved in the initial stages of carcinogenesis [46–49]. Our data also confirmed the involvement of the estrogen signaling pathway in maternal malnutrition inducing early life prostate carcinogenesis in rat offspring. Previous experimental evidence has demonstrated that early life exposure to exogenous estrogenic compounds, such as BPA and Phthalates, may epigenetically reprogram prostate developmental biology, lead to prostate carcinogenesis with aging [16, 50].
Protein-protein interactions play a crucial role in the control of cellular functions, signal transduction, and metabolism; as such, understanding these interactions may help us to identify molecular mechanisms involved in maternal malnutrition-induced prostatic disorders. The RAB family of proteins, identified by a PPI network analysis, belongs to the RAS (rat sarcoma) superfamily of small GTPase. RAB comprises a family of 66 members (the number of RAB-GTPases is conserved from yeast to humans) [51, 52], which function as molecular regulators essential for the localization and function of the membrane and secretory proteins such as hormones, growth factors, and their membrane receptors. As such, RAB activates several signaling pathways, including the MAPK pathway involved in cell growth and proliferation [53] and the PI3K/AKT/mTOR pathway that stimulates protein synthesis, cell growth, and inhibits apoptosis [54]. Altered expression and the activity of RAB members have been implicated in the development of several disorders, ranging from neurological disorders to diabetes [55]. Aberrant expression of RAB proteins has also been described in multiple cancers, such as lung, brain, and breast. RAB 35, which appeared to be deregulated in our study, can act as an oncogene [56]. The GTPase-deficient RAB35 mutant (RAB35Q67L) activates the PI3K signaling pathway independently of growth factor stimulation and suppresses apoptosis in human embryonic kidney HEK293E cells [57]. The cluster formed CALR, HSPA5, P4HB and, PDIA6 is a potential indicator of maternal malnutrition on the endoplasmic reticulum dysfunction in the prostate of offspring since they act as the fundamental molecular machinery for correct protein folding and Ca2+ homeostasis. In a mouse model of caloric restriction (CR), Schafer et al. [58] compared the influence of CR on the hippocampus at younger-adult and older-adult time points and identified the upregulation of HSPA1B, HSPA5, PDIA4, PDIA6, and CALR. Other authors have also associated the deregulation of these proteins in several types of cancer, including breast carcinoma, hepatoma cells, non-small cell lung cancer, and glioma [59–63]. Although epigenetic modifications of histones, such as histone lysine methylation and demethylation, histone lysine acetylation and deacetylation have been implicated in the modulation of gene expression in the physiological and pathological conditions [64, 65], deregulation of histone expression itself has also been described in several types of malignancies. Xie et al. [66] demonstrated upregulation of hub genes formed by HIST1H1B, HIST1H2AJ, HIST1H2AM, HIST1H2BI, HIST1H2BO, HIST1H3B, HIST1H3F, HIST1H3H, HIST1H4C, and HIST1H4D in breast cancer, indicating that higher expression of these histones was associated with poor overall survival, relapse-free survival, and distant metastasis-free survival. Interestingly, we also observed an increased expression of HIST1 gene members (HIST1H2AA, HIST1H2AH, HIST1H2AK, HIST1H2AN) in the VP of both young and older rats exposed to maternal LPD. This result highlights the potential involvement of the upregulation of histone proteins on prostatic disorders.
Another cluster is formed by enzymes acting in response to oxidative stress as GST, PRDX, and TXN1. The superfamily of GSTs acts in several mechanisms of cellular detoxification, resistance to anticancer drugs, pollutants, and chemicals [67], and the overexpression of GSTs is also associated with the presence of an inflammatory process [68]. It has been demonstrated that low expression of GSTs increases reactive oxygen species in spermatozoids, leading to a degradation of the plasma membrane and a loss of sperm viability [69–71]. In the prostate gland, GSTs are mainly expressed in the basal cells [72], and their overexpression is associated with epithelial disorders [73], DNA oxidation, and methylation [74, 75]. PRDX-5 is known to act as a redox sensor in the cytosol and several cellular compartments, and silencing it makes the cell more susceptible to DNA damage and apoptosis [76]. In gastric cancer, overexpression of PRDX5 alters the epithelial to mesenchymal transition (EMT) mechanism, with a poor prognosis for patients [77] being correlated. TXN recycles oxidized PRDXs, and this function is essential to balance intracellular oxidative stress [78, 79]. The increased expression of TXN in prostate tissue has been positively correlated with the progression of Gleason score in patients with PCa [80], indicating that transformed cells express higher levels of Trx 1 compared with normal cells. On the other hand, the treatment of prostate cancer cells with natural bioactive compounds reduces TXN expression, collaborating with the apoptosis of these cells [81]. Thus, the high expression of TXN in the prostate of maternal LPD offspring could be related to the development of carcinoma in situ, as observed in older rats.
Considering that groups of interacting proteins are deregulated in both younger and older undernourished offspring, it is possible that histones and RAS-GTPase families and proteins related to oxidative stress, besides those involved with endoplasmic reticulum metabolism and function, may participate in the long-term effect of maternal malnutrition on the prostate of offspring. These results become more relevant with the identification of several of these proteins in patients and mice model of PCa by in silico analysis.
Conclusions
In the present study, we show that maternal exposure to low protein diet deregulated molecular pathways involved in prostate development early in life, which may act as risk factors for prostate carcinogenesis with aging. Estrogenic signaling pathways, endoplasmic reticulum functions related to detoxification, energy metabolism, and molecular sensors of protein folding and Ca2+ homeostasis, besides histone, and RAS-GTPase family of proteins appear to be involved in this process. Knowledge of these factors may raise discussions regarding the role of maternal dietary intervention as a favorable public policy for the lifelong prevention of chronic diseases.
Materials and Methods
Animals and experimental design
The detailed experimental design is described by Santos et al. [16]. Briefly, after the determination of pregnancy on gestational day 1 (GD1), pregnant rats were distributed into two experimental groups (n=6/group): Control (CTR): dams fed a normal protein diet (17% protein) and gestational and lactational low protein (GLLP): dams fed a low protein diet (LPD) during gestational and lactational periods. Normal and LPD diets were provided by PragSoluções (PragSoluções, SP, Brazil). All diets were isocaloric and normosodic (Supplementary Table 1). The male offspring were euthanized on a postnatal day (PND) 21 (weaning) (n=12/group) and PND 540 (n=12/group). The offspring, which were euthanized on PND 540, had free access to a normal protein diet after weaning until the end of the experiment. The animals were euthanized by an overdose of anesthesia (ketamine/xylazine) followed by decapitation, weighing, and the blood and ventral prostate (VP) were collected and processed by a different analysis as described below. The body weight, and VP weight, and hormonal levels were analyzed using a Student t-test, and statistical differences were considered when p < 0.05. The animal procedures were approved by the Biosciences Institute/UNESP Ethics Committee for Animal Experimentation (Protocol #573) following the ethical animal research principles and the Brazilian legislation established by the Brazilian Council of Control in Animal Experimentation.
Hormone analysis
Blood samples from offspring (n=12/group) were centrifuged (2400 g for 20 minutes), and sera were used to determine the concentrations of estrogen (17β-estradiol, Monobind®, 4925-300 CA, USA sensitivity: 6.5 pg/mL) and testosterone (17β-hydroxy-4-androstene-3-one, Monobind®, 3725-300A, CA. sensitivity: 0.038 ng/mL). The hormonal qualifications were determined in 96-well plates using the ELISA plate reader (Epoch™, Biotek Instruments, VT, USA) following the manufacturers' protocol.
Selection of prostate samples for mass spectrometry analysis
In a previous study, Santos et al. [16] demonstrated that maternal exposure to LPD induced a delay in prostatic growth on PND 21, which was associated with prostate carcinogenesis in older rats on PND 540. Slides of the left VP lobes (n=3/group) were stained with hematoxylin-eosin (HE) and analyzed using a Leica DMLB 80 microscope To exemplify the histological characteristics of the VP lobes from the CTR and GLLP groups on PND 21 and 540. Based on these results, the contralateral right VP lobes (n=3/group) from each group were submitted to mass spectrometry.
Immunohistochemistry
Histological sections of 5 μm (n = 6 per group) were processed as described by Santos et al. [16]. After the initial steps, the slides were boiled for 30 min in 10 mM sodium citrate solution (pH 6.0) for antigen retrieval. Prostatic sections were blocked in 5% nonfat milk diluted in phosphate-buffered saline (PBS) and incubated with anti-Calreticulin antibody (ab2908) specific primary antibody overnight at 4°C. Slides were washed in PBS and incubated for one hour at room temperature in horseradish peroxidase (HRP)-conjugated secondary antibody. The slides were washed, and the reaction was developed using 3,3′-Diaminobenzidine (DAB, Sigma) and counterstained with hematoxylin for 30 seconds. The reactions were analyzed using a Leica DMLB 80 microscope.
RT-qPCR
Prostate samples (n= 6 per group) from all experimental groups on PND 21 and PND 540 were used to total RNA extraction using TRIzol® Reagent (ThermoFisher aScientific) according to the manufacturer's recommendations. RNA integrity was evaluated by capillary electrophoresis using a 2100 Bioanalyzer (Agilent). Only samples with an RNA integrity number (RIN) ≥ 7.0 were used. The extracted RNA was treated with DNase I (Amplification Grade; ThermoFisher Scientific). The synthesis of cDNA was performed using a High Capacity cDNA Archive Kit (ThermoFisher Scientific) according to the manufacturer's guidelines. Expression levels of CALR mRNA were measured by RT-qPCR using the QuantStudio™ 12K Flex Real-Time PCR System (Thermo Fisher Scientific). All qPCRs performed were compliant with the Minimum Information for Publication of Quantitative Real-Time PCR experiments (MIQE) guidelines [82]. The cDNA samples were amplified using SYBR® Green Master Mix (ThermoFisher Scientific), and specific primers were synthesized by Invitrogen to the CALR gene, forward: GCCAGACACTGGTGGTACAGTTC reverse: CGCCCCCACAGTCGATATT. Relative quantification of expression was performed by the 2−ΔΔCt method [83] using DataAssistTM v3.01 software (Thermo Fisher Scientific). According to the expression stability among all samples, the reference gene GUSB (β-glucuronidase) and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) were used to normalize mRNA expression.
Mass spectrometry
The mass spectrometry protocol was based on a previous study by Gabriel Kuniyoshi et al. [84], Dionizio et al. [85], and Da Silva-Gomes et al. [86], with modifications. Briefly, protein extraction was carried out by homogenizing three VPs lobes from each experimental group on PND 21 and 540 in extraction buffer containing 0.01 M Tris-HCl, 0.005 M phenylmethylsulfonyl fluoride, 1% protease inhibitor, 0.065 M dithiothreitol, 8 M urea (in a proportion of 30mg tissue/100 μg buffer). The homogenate was vortexed for 2-3 min and centrifuged for 15 min at 9,690 g and 4°C. The supernatant was recovered, and the total protein was quantified by Bradford assay using the BSA standard [87]. Samples were grouped to constitute three pools of 50 μg proteins each in a total of 50 μL (1 μg/μL). Next, the samples were incubated for 60 min at 37°C with 10 μL of 50 mM ammonium bicarbonate and 25 μl of 0.2% surfactant solution, followed by incubation with 2.5 μL of 0.1 M dithiothreitol for 40 min at 37°C. Carbamidomethylation was performed with 2.5 μL of 0.3 M iodoacetamide, incubated for 30 min at room temperature, and protected from light. Then, samples were subjected to proteolytic digestion overnight at 37°C using 0.05 μg/μL trypsin diluted in 0.05 M Ammonium bicarbonate, followed by incubation with 10 μL trifluoroacetic acid 5% for 90 min at 37°C. The samples were centrifuged at 14,000 RPM 4ºC for 30 minutes. After this step, the samples were desalted using Sep-Pak Vac C18 (Waters Manchester, UK) columns, reduced in a concentrator, and maintained at -20°C until the time of analysis by mass spectrometry.
The analysis of the tryptic peptide was performed using the nanoACQUITY UPLC system (Waters, Manchester, UK) coupled to a Xevo Q-TOF G2 mass spectrometer (Waters, Manchester, UK) equipped with nanoACQUITY HSS T3, analytical reverse-phase column (75μmX150 mm, 1.8μm particle size, Waters) previously equilibrated with 7% of mobile phase B (100% ACN + 0.1% formic acid). The peptides were separated by a linear gradient of 7-85% mobile phase B for 70 min at a flow rate of 0,35 μL/min, and the column temperature was maintained at 45°C. The MS was operated in positive ion mode, with a data acquisition time of 75 min. The data obtained were processed using the software Protein Lynx Global Server (PLGS) version 3.03 (Waters Co., Manchester, UK). Protein identification was obtained using an ion count algorithm incorporated into the software. The data obtained were searched in the database of the species Rattus norvegicus downloaded from the UniProt catalog (Universal Protein Resource) in December 2017 (https://www.uniprot.org/). Differentially expressed proteins (DEP) between experimental groups were obtained using PLGS software, considering p < 0.05 for downregulated proteins and p > 0.95 for upregulated proteins.
Functional annotation analysis
KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/) was used to determine the enrichment pathways related to our DEP in the KEGG (https://www.genome.jp/kegg/) and PANTHER (http://pantherdb.org) databases. The cut-off criterion used was an adjusted p-value < 0.05. Also, we used the Ligand Perturbation database from the Enrichr tool (http://amp.pharm.mssm.edu/Enrichr) to compare the set of DEP from those extracted from GEO comparing human or mouse cells before and after treatment with endogenous ligands. We used the top 10 most enriched terms with a p-value < 0.05 [88]. The STRING tool (http://string-db.org/) was used to construct the protein-protein interaction (PPI) network associated with our DEP by searching neighbor interactors with our imputed proteins. To avoid false positive interactions, we selected a high confidence score (0.7), associated with experiments and a database as two stringent evidence channels [89].
Relevance of deregulated proteins in human and mouse model of PCa: in silico validation
To give further insights into the relationship between maternal malnutrition and PCa, we compared our set of DEP with RNA-seq data from a transgenic mice model for PCa: PB-Cre/PtenloxP/loxP. In this study, Jurmeister et al. [90] described data from the RNA-seq of four prostate lobes (ventral, anterior, dorsal, and lateral) at different stages of tumorigenesis: low-grade prostate intraepithelial neoplasia (PIN), medium-stage tumors (Medium) and advanced-stage tumors (Advanced). The dataset was downloaded from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE94574. We considered differentially expressed genes: < -1.3 Log2FC > 1.3, adjusted p-value < 0.05. We also identified differentially expressed genes between normal from Genotype-Tissue Expression (GTEx) with 221 patients/samples and PCa human samples extracted from RNA-seq data using Prostate Adenocarcinoma (TCGA, PanCancer Atlas) with 488 patients/samples data analyzed using the GEPIA database (Gene Expression Profiling Interactive Analysis) (http://gepia.cancer-pku.cn/) [91]. We considered differently expressed genes with < -1 Log2FC > 1 and q-value < 0.05. This set of genes was compared with our DEP to identify possible molecular mechanisms shared by our samples and those from human PCa. Additionally, the commonly upregulated proteins identified in our sample and RNA-seq of prostatic tumor samples identified by GEPIA were submitted to The Human Protein Atlas (HPA) (https://www.proteinatlas.org/) database to demonstrate the distribution and localization of these proteins in normal and tumor samples by immunohistochemistry.
Abbreviations
CALR: Calreticulin;
CR: Caloric restriction;
CTR: Control group;
DAB: 3,3′-Diaminobenzidine;
DEP: Differentially expressed proteins;
DOHaD: developmental origins of health and disease;
EGF: Epidermal growth factor;
EMT: Epithelial to mesenchymal transition;
FGF: Fibroblast growth factor;
FOAD: Fetal Origin of Adult Diseases;
GAPDH: Glyceraldehyde 3-phosphate dehydrogenase;
GD: Gestational day;
GEPIA: Gene Expression Profiling Interactive Analysis;
GLLP: Gestational and lactational low protein group;
GLOBOCAN: Global Cancer Statistics;
GSTM2: Glutathione S-transferase Mu 2;
GTEx: Genotype-Tissue Expression;
GUSB: β-glucuronidase;
H2AFJ: Histone H2A.J;
HBA1: Hemoglobin Subunit Alpha 1;
HE: Hematoxylin–eosin;
HPA: Human Protein Atlas;
HSPA5: Heat shock protein family A member 5;
LPD: Low Protein Diet;
MAPK: Mitogen-activated protein kinase;
MM: Maternal malnutrition;
P4HB: Protein disulfide isomerase-4;
PBS: Phosphate-buffered saline;
PCa: Prostate cancer;
PDIA6: Protein Disulfide Isomerase Family A Member 6;
PI3K: Phosphatidylinositol 3'-kinase;
PIN: Intraepithelial neoplasia;
PND: Post-natal day;
PPI: Protein-Protein interaction;
PRDX5: Peroxiredoxin-5;
RAB: Ras-related protein;
RAP1: Ras-proximate-1;
RAS: Rat sarcoma;
RIN: RNA integrity number;
TCGA: The Cancer Genomic Atlas;
TXN1: Thioredoxin 1;
UniProt: Universal Protein Resource;
VP: Ventral prostate.
Author Contributions
Conceptualization, S.A.A.S., and L.A.J.; methodology, S.A.A.S., A.C.L.C., F.B.C., KT.C., M.N.F, J.C.S.V., P.G.P., L.M.F.P., M.B, L.A.J.; formal analysis, S.A.A.S., L.A.J.; investigation, S.A.A.S., A.C.C., F.B.C., L.A.J.; resources, S.A.A.S., A.C.L.C., L.A.J; data curation, S.A.A.S., S.L.F., L.A.J.; writing—original draft preparation, S.A.A.S., A.C.L.C., F.B.C., L.A.J.; writing—review and editing, all authors; supervision, L.A.J; project administration, L.A.J.; funding acquisition, L.A.J.
Acknowledgments
We thank Wellerson Scarano (Institute of Biosciences of Botucatu, UNESP) for helping with histopathological and bioinformatics analyses.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
This research was funded by São Paulo Research Foundation (FAPESP, grant 2017/01063-7, LAJ). The following grants also supported this work: FAPESP (grant # 2017/08715-0, ACLC and 2014/08531-8, SAAS); National Council for Scientific and Technological Development (CNPq grant # 310663/ 2018- 0); “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES)” - Finance Code 001 (scholarship to A.C.L.C).
References
-
1.
Chatterji S, Byles J, Cutler D, Seeman T, Verdes E. Health, functioning, and disability in older adults—present status and future implications. Lancet. 2015; 385:563–75. https://doi.org/10.1016/S0140-6736(14)61462-8 [PubMed]
-
2.
Newman CB, Preiss D, Tobert JA, Jacobson TA, Page RL 2n, Goldstein LB, Chin C, Tannock LR, Miller M, Raghuveer G, Duell PB, Brinton EA, Pollak A, et al, and American Heart Association Clinical Lipidology, Lipoprotein, Metabolism and Thrombosis Committee, a Joint Committee of the Council on Atherosclerosis, Thrombosis and Vascular Biology and Council on Lifestyle and Cardiometabolic Health; Council on Cardiovascular Disease in the Young; Council on Clinical Cardiology; and Stroke Council. Statin safety and associated adverse events: a scientific statement from the American heart association. Arterioscler Thromb Vasc Biol. 2019; 39:e38–81. https://doi.org/10.1161/ATV.0000000000000073 [PubMed]
-
3.
Barker DJ, Osmond C, Golding J, Kuh D, Wadsworth ME. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ. 1989; 298:564–67. https://doi.org/10.1136/bmj.298.6673.564 [PubMed]
-
4.
Dickinson FM, Pyone T, van den Broek N. Experiences from the field: maternal, reproductive and child health data collection in humanitarian and emergency situations. Int Health. 2016; 8:83–88. https://doi.org/10.1093/inthealth/ihv045 [PubMed]
-
5.
McKerracher L, Moffat T, Barker M, McConnell M, Atkinson SA, Murray-Davis B, McDonald SD, Sloboda DM. Knowledge about the developmental origins of health and disease is independently associated with variation in diet quality during pregnancy. Matern Child Nutr. 2020; 16:e12891. https://doi.org/10.1111/mcn.12891 [PubMed]
-
6.
Barker DJ, Bull AR, Osmond C, Simmonds SJ. Fetal and placental size and risk of hypertension in adult life. BMJ. 1990; 301:259–62. https://doi.org/10.1136/bmj.301.6746.259 [PubMed]
-
7.
Wadhwa PD, Buss C, Entringer S, Swanson JM. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med. 2009; 27:358–68. https://doi.org/10.1055/s-0029-1237424 [PubMed]
-
8.
Schulz LC. The dutch hunger winter and the developmental origins of health and disease. Proc Natl Acad Sci USA. 2010; 107:16757–58. https://doi.org/10.1073/pnas.1012911107 [PubMed]
-
9.
Lumey LH, Van Poppel FW. The dutch famine of 1944-45: mortality and morbidity in past and present generations. Soc Hist Med. 1994; 7:229–46. https://doi.org/10.1093/shm/7.2.229 [PubMed]
-
10.
Hoek HW, Susser E, Buck KA, Lumey LH, Lin SP, Gorman JM. Schizoid personality disorder after prenatal exposure to famine. Am J Psychiatry. 1996; 153:1637–39. https://doi.org/10.1176/ajp.153.12.1637 [PubMed]
-
11.
Kahn HS, Stein AD, Lumey LH. Prenatal environmental exposures that may influence β-cell function or insulin sensitivity in middle age. J Dev Orig Health Dis. 2010; 1:300–09. https://doi.org/10.1017/S2040174410000474 [PubMed]
-
12.
Walker CL, Ho SM. Developmental reprogramming of cancer susceptibility. Nat Rev Cancer. 2012; 12:479–86. https://doi.org/10.1038/nrc3220 [PubMed]
-
13.
Keinan-Boker L, Vin-Raviv N, Liphshitz I, Linn S, Barchana M. Cancer incidence in Israeli jewish survivors of world war II. J Natl Cancer Inst. 2009; 101:1489–500. https://doi.org/10.1093/jnci/djp327 [PubMed]
-
14.
Barker DJ, Osmond C, Thornburg KL, Kajantie E, Eriksson JG. A possible link between the pubertal growth of girls and prostate cancer in their sons. Am J Hum Biol. 2012; 24:406–10. https://doi.org/10.1002/ajhb.22222 [PubMed]
-
15.
Varuzza MB, Zapaterini JR, Colombelli KT, Barquilha CN, Justulin LA Jr, Muñoz-de-Toro M, Kass L, Barbisan LF. Impact of gestational low protein diet and postnatal bisphenol a exposure on chemically induced mammary carcinogenesis in female offspring rats. Environ Toxicol. 2019; 34:1263–72. https://doi.org/10.1002/tox.22827 [PubMed]
-
16.
Santos SA, Camargo AC, Constantino FB, Colombelli KT, Mani F, Rinaldi JC, Franco S, Portela LM, Duran BO, Scarano WR, Hinton BT, Felisbino SL, Justulin LA. Maternal low-protein diet impairs prostate growth in young rat offspring and induces prostate carcinogenesis with aging. J Gerontol A Biol Sci Med Sci. 2019; 74:751–59. https://doi.org/10.1093/gerona/gly118 [PubMed]
-
17.
Scarano WR, Bedrat A, Alonso-Costa LG, Aquino AM, Fantinatti B, Justulin LA, Barbisan LF, Freire PP, Flaws JA, Bernardo L. Exposure to an environmentally relevant phthalate mixture during prostate development induces microRNA upregulation and transcriptome modulation in rats. Toxicol Sci. 2019; 171:84–97. https://doi.org/10.1093/toxsci/kfz141 [PubMed]
-
18.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424. https://doi.org/10.3322/caac.21492 [PubMed]
-
19.
Gardner WA. Hypothesis: the prenatal origins of prostate cancer. Hum Pathol. 1995; 26:1291–92. https://doi.org/10.1016/0046-8177(95)90291-0 [PubMed]
-
20.
Powell IJ, Meyskens FL Jr. African American men and hereditary/familial prostate cancer: intermediate-risk populations for chemoprevention trials. Urology. 2001; 57:178–81. https://doi.org/10.1016/s0090-4295(00)00968-7 [PubMed]
-
21.
Henderson BE, Bernstein L, Ross RK, Depue RH, Judd HL. The early in utero oestrogen and testosterone environment of blacks and whites: potential effects on male offspring. Br J Cancer. 1988; 57:216–18. https://doi.org/10.1038/bjc.1988.46 [PubMed]
-
22.
Elias SG, Peeters PH, Grobbee DE, van Noord PA. Breast cancer risk after caloric restriction during the 1944-1945 dutch famine. J Natl Cancer Inst. 2004; 96:539–46. https://doi.org/10.1093/jnci/djh087 [PubMed]
-
23.
Dirx MJ, van den Brandt PA, Goldbohm RA, Lumey LH. Energy restriction in childhood and adolescence and risk of prostate cancer: results from the Netherlands cohort study. Am J Epidemiol. 2001; 154:530–37. https://doi.org/10.1093/aje/154.6.530 [PubMed]
-
24.
Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol a increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006; 66:5624–32. https://doi.org/10.1158/0008-5472.CAN-06-0516 [PubMed]
-
25.
Prins GS, Hu WY, Xie L, Shi GB, Hu DP, Birch L, Bosland MC. Evaluation of bisphenol a (BPA) exposures on prostate stem cell homeostasis and prostate cancer risk in the NCTR-sprague-dawley rat: an NIEHS/FDA CLARITY-BPA consortium study. Environ Health Perspect. 2018; 126:117001. https://doi.org/10.1289/EHP3953 [PubMed]
-
26.
Prins GS, Ye SH, Birch L, Zhang X, Cheong A, Lin H, Calderon-Gierszal E, Groen J, Hu WY, Ho SM, van Breemen RB. Prostate cancer risk and DNA methylation signatures in aging rats following developmental BPA exposure: a dose-response analysis. Environ Health Perspect. 2017; 125:077007. https://doi.org/10.1289/EHP1050 [PubMed]
-
27.
Cheong JN, Wlodek ME, Moritz KM, Cuffe JS. Programming of maternal and offspring disease: impact of growth restriction, fetal sex and transmission across generations. J Physiol. 2016; 594:4727–40. https://doi.org/10.1113/JP271745 [PubMed]
-
28.
Trichopoulos D, MacMahon B, Brown J. Socioeconomic status, urine estrogens, and breast cancer risk. J Natl Cancer Inst. 1980; 64:753–55. [PubMed]
-
29.
Justulin LA, Dos Santos SAA, Damasceno DC, Scarano WR, Felisbino SL. Chapter 20: Nutrition and prostate cancer prevention. Food Chemistry, Function and Analysis. Royal Society of Chemistry; 2020; 21:392–411. https://doi.org/10.1039/9781788016506-00392
-
30.
de Mello Santos T, Cavariani MM, Pereira DN, Schimming BC, Chuffa LG, Domeniconi RF. Maternal protein restriction modulates angiogenesis and AQP9 expression leading to a delay in postnatal epididymal development in rat. Cells. 2019; 8:1094. https://doi.org/10.3390/cells8091094 [PubMed]
-
31.
Barbeito-Andrés J, Pezzuto P, Higa LM, Dias AA, Vasconcelos JM, Santos TM, Ferreira JC, Ferreira RO, Dutra FF, Rossi AD, Barbosa RV, Amorim CK, De Souza MP, et al. Congenital zika syndrome is associated with maternal protein malnutrition. Sci Adv. 2020; 6:eaaw6284. https://doi.org/10.1126/sciadv.aaw6284 [PubMed]
-
32.
Colombelli KT, Santos SA, Camargo AC, Constantino FB, Barquilha CN, Rinaldi JC, Felisbino SL, Justulin LA. Impairment of microvascular angiogenesis is associated with delay in prostatic development in rat offspring of maternal protein malnutrition. Gen Comp Endocrinol. 2017; 246:258–69. https://doi.org/10.1016/j.ygcen.2016.12.016 [PubMed]
-
33.
Kim HG, Kassis J, Souto JC, Turner T, Wells A. EGF receptor signaling in prostate morphogenesis and tumorigenesis. Histol Histopathol. 1999; 14:1175–82. https://doi.org/10.14670/HH-14.1175 [PubMed]
-
34.
Donjacour AA, Thomson AA, Cunha GR. FGF-10 plays an essential role in the growth of the fetal prostate. Dev Biol. 2003; 261:39–54. https://doi.org/10.1016/s0012-1606(03)00250-1 [PubMed]
-
35.
Ma Y, Kakudo N, Morimoto N, Lai F, Taketani S, Kusumoto K. Fibroblast growth factor-2 stimulates proliferation of human adipose-derived stem cells via src activation. Stem Cell Res Ther. 2019; 10:350. https://doi.org/10.1186/s13287-019-1462-z [PubMed]
-
36.
Thomson AA, Cunha GR. Prostatic growth and development are regulated by FGF10. Development. 1999; 126:3693–701. [PubMed]
-
37.
Pillai A, Patel S, Ranadive I, Desai I, Balakrishnan S. Fibroblast growth factor-2 signaling modulates matrix reorganization and cell cycle turnover rate in the regenerating tail of hemidactylus flaviviridis. Acta Histochem. 2020; 122:151464. https://doi.org/10.1016/j.acthis.2019.151464 [PubMed]
-
38.
Ropiquet F, Giri D, Lamb DJ, Ittmann M. FGF7 and FGF2 are increased in benign prostatic hyperplasia and are associated with increased proliferation. J Urol. 1999; 162:595–99. [PubMed]
-
39.
Meng Z, Moroishi T, Guan KL. Mechanisms of hippo pathway regulation. Genes Dev. 2016; 30:1–17. https://doi.org/10.1101/gad.274027.115 [PubMed]
-
40.
Hong AW, Meng Z, Guan KL. The hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol. 2016; 13:324–37. https://doi.org/10.1038/nrgastro.2016.59 [PubMed]
-
41.
Zacharopoulou N, Tsapara A, Kallergi G, Schmid E, Tsichlis PN, Kampranis SC, Stournaras C. The epigenetic factor KDM2B regulates cell adhesion, small rho GTPases, actin cytoskeleton and migration in prostate cancer cells. Biochim Biophys Acta Mol Cell Res. 2018; 1865:587–97. https://doi.org/10.1016/j.bbamcr.2018.01.009 [PubMed]
-
42.
Mason MD, Davies G, Jiang WG. Cell adhesion molecules and adhesion abnormalities in prostate cancer. Crit Rev Oncol Hematol. 2002; 41:11–28. https://doi.org/10.1016/s1040-8428(01)00171-8 [PubMed]
-
43.
Albany C, Hahn NM. Heat shock and other apoptosis-related proteins as therapeutic targets in prostate cancer. Asian J Androl. 2014; 16:359–63. https://doi.org/10.4103/1008-682X.126400 [PubMed]
-
44.
Giampietri C, Petrungaro S, Conti S, Facchiano A, Filippini A, Ziparo E. Cancer microenvironment and endoplasmic reticulum stress response. Mediators Inflamm. 2015; 2015:417281. https://doi.org/10.1155/2015/417281 [PubMed]
-
45.
Jaud M, Philippe C, Di Bella D, Tang W, Pyronnet S, Laurell H, Mazzolini L, Rouault-Pierre K, Touriol C. Translational regulations in response to endoplasmic reticulum stress in cancers. Cells. 2020; 9:540. https://doi.org/10.3390/cells9030540 [PubMed]
-
46.
Knudsen BS, Vasioukhin V. Mechanisms of prostate cancer initiation and progression. Adv Cancer Res. 2010; 109:1–50. https://doi.org/10.1016/B978-0-12-380890-5.00001-6 [PubMed]
-
47.
Shiao SL, Chu GC, Chung LW. Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett. 2016; 380:340–48. https://doi.org/10.1016/j.canlet.2015.12.022 [PubMed]
-
48.
Humphrey PA. Histopathology of prostate cancer. Cold Spring Harb Perspect Med. 2017; 7:a030411. https://doi.org/10.1101/cshperspect.a030411 [PubMed]
-
49.
Hütz K, Zeiler J, Sachs L, Ormanns S, Spindler V. Loss of desmoglein 2 promotes tumorigenic behavior in pancreatic cancer cells. Mol Carcinog. 2017; 56:1884–95. https://doi.org/10.1002/mc.22644 [PubMed]
-
50.
Prins GS, Ho SM. Early-life estrogens and prostate cancer in an animal model. J Dev Orig Health Dis. 2010; 1:365–70. https://doi.org/10.1017/S2040174410000577 [PubMed]
-
51.
Stenmark H, Olkkonen VM. The rab GTPase family. Genome Biol. 2001; 2:REVIEWS3007. https://doi.org/10.1186/gb-2001-2-5-reviews3007 [PubMed]
-
52.
Li G, Marlin MC. Rab family of GTPases. Methods Mol Biol. 2015; 1298:1–15. https://doi.org/10.1007/978-1-4939-2569-8_1 [PubMed]
-
53.
Nan X, Tamgüney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, Chu S. ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc Natl Acad Sci USA. 2015; 112:7996–8001. https://doi.org/10.1073/pnas.1509123112 [PubMed]
-
54.
Castellano E, Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011; 2:261–74. https://doi.org/10.1177/1947601911408079 [PubMed]
-
55.
Abd Elmageed ZY, Yang Y, Thomas R, Ranjan M, Mondal D, Moroz K, Fang Z, Rezk BM, Moparty K, Sikka SC, Sartor O, Abdel-Mageed AB. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014; 32:983–97. https://doi.org/10.1002/stem.1619 [PubMed]
-
56.
Gopal Krishnan PD, Golden E, Woodward EA, Pavlos NJ, Blancafort P. Rab GTPases: emerging oncogenes and tumor suppressive regulators for the editing of survival pathways in cancer. Cancers (Basel). 2020; 12:259. https://doi.org/10.3390/cancers12020259 [PubMed]
-
57.
Wheeler DB, Zoncu R, Root DE, Sabatini DM, Sawyers CL. Identification of an oncogenic RAB protein. Science. 2015; 350:211–17. https://doi.org/10.1126/science.aaa4903 [PubMed]
-
58.
Schafer MJ, Dolgalev I, Alldred MJ, Heguy A, Ginsberg SD. Calorie restriction suppresses age-dependent hippocampal transcriptional signatures. PLoS One. 2015; 10:e0133923. https://doi.org/10.1371/journal.pone.0133923 [PubMed]
-
59.
Bini L, Magi B, Marzocchi B, Arcuri F, Tripodi S, Cintorino M, Sanchez JC, Frutiger S, Hughes G, Pallini V, Hochstrasser DF, Tosi P. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 1997; 18:2832–41. https://doi.org/10.1002/elps.1150181519 [PubMed]
-
60.
Bai Y, Wang G, Fu W, Lu Y, Wei W, Chen W, Wu X, Meng H, Feng Y, Liu Y, Li G, Wang S, Wang K, et al. Circulating essential metals and lung cancer: Risk assessment and potential molecular effects. Environ Int. 2019; 127:685–693. https://doi.org/10.1016/j.envint.2019.04.021 [PubMed]
-
61.
Yu LR, Zeng R, Shao XX, Wang N, Xu YH, Xia QC. Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis. 2000; 21:3058–68. https://doi.org/10.1002/1522-2683(20000801)21:14<3058::AID-ELPS3058>3.0.CO;2-U [PubMed]
-
62.
Zou H, Wen C, Peng Z, Shao YΥ, Hu L, Li S, Li C, Zhou HH. P4HB and PDIA3 are associated with tumor progression and therapeutic outcome of diffuse gliomas. Oncol Rep. 2018; 39:501–10. https://doi.org/10.3892/or.2017.6134 [PubMed]
-
63.
Albakova Z, Armeev GA, Kanevskiy LM, Kovalenko EI, Sapozhnikov AM. HSP70 multi-functionality in cancer. Cells. 2020; 9:587. https://doi.org/10.3390/cells9030587 [PubMed]
-
64.
Shanmugam MK, Arfuso F, Arumugam S, Chinnathambi A, Jinsong B, Warrier S, Wang LZ, Kumar AP, Ahn KS, Sethi G, Lakshmanan M. Role of novel histone modifications in cancer. Oncotarget. 2017; 9:11414–26. https://doi.org/10.18632/oncotarget.23356 [PubMed]
-
65.
Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016; 8:a019521. https://doi.org/10.1101/cshperspect.a019521 [PubMed]
-
66.
Xie W, Zhang J, Zhong P, Qin S, Zhang H, Fan X, Yin Y, Liang R, Han Y, Liao Y, Yu X, Long H, Lv Z, et al. Expression and potential prognostic value of histone family gene signature in breast cancer. Exp Ther Med. 2019; 18:4893–903. https://doi.org/10.3892/etm.2019.8131 [PubMed]
-
67.
Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005; 45:51–88. https://doi.org/10.1146/annurev.pharmtox.45.120403.095857 [PubMed]
-
68.
Bogdani M, Henschel AM, Kansra S, Fuller JM, Geoffrey R, Jia S, Kaldunski ML, Pavletich S, Prosser S, Chen YG, Lernmark A, Hessner MJ. Biobreeding rat islets exhibit reduced antioxidative defense and n-acetyl cysteine treatment delays type 1 diabetes. J Endocrinol. 2013; 216:111–23. https://doi.org/10.1530/JOE-12-0385 [PubMed]
-
69.
Llavanera M, Mateo-Otero Y, Bonet S, Barranco I, Fernández-Fuertes B, Yeste M. The triple role of glutathione s-transferases in mammalian male fertility. Cell Mol Life Sci. 2020; 77:2331–42. https://doi.org/10.1007/s00018-019-03405-w [PubMed]
-
70.
Fafula RV, Paranyak NM, Besedina AS, Vorobets DZ, Iefremova UP, Onufrovych OK, Vorobets ZD. Biological significance of glutathione s-transferases in human sperm cells. J Hum Reprod Sci. 2019; 12:24–28. https://doi.org/10.4103/jhrs.JHRS_106_18 [PubMed]
-
71.
Kan HP, Wu FL, Guo WB, Wang YZ, Li JP, Huang YQ, Li SG, Liu JP. Null genotypes of GSTM1 and GSTT1 contribute to male factor infertility risk: a meta-analysis. Fertil Steril. 2013; 99:690–96. https://doi.org/10.1016/j.fertnstert.2012.10.037 [PubMed]
-
72.
De Marzo AM, Meeker AK, Zha S, Luo J, Nakayama M, Platz EA, Isaacs WB, Nelson WG. Human prostate cancer precursors and pathobiology. Urology. 2003; 62:55–62. https://doi.org/10.1016/j.urology.2003.09.053 [PubMed]
-
73.
Sanches LD, Santos SA, Carvalho JR, Jeronimo GD, Favaro WJ, Reis MD, Felisbino SL, Justulin LA Jr. Protective effect of γ-tocopherol-enriched diet on N-methyl-N-nitrosourea-induced epithelial dysplasia in rat ventral prostate. Int J Exp Pathol. 2013; 94:362–72. https://doi.org/10.1111/iep.12042 [PubMed]
-
74.
Martignano F, Gurioli G, Salvi S, Calistri D, Costantini M, Gunelli R, De Giorgi U, Foca F, Casadio V. GSTP1 methylation and protein expression in prostate cancer: diagnostic implications. Dis Markers. 2016; 2016:4358292. https://doi.org/10.1155/2016/4358292 [PubMed]
-
75.
Schnekenburger M, Karius T, Diederich M. Regulation of epigenetic traits of the glutathione s-transferase P1 gene: from detoxification toward cancer prevention and diagnosis. Front Pharmacol. 2014; 5:170. https://doi.org/10.3389/fphar.2014.00170 [PubMed]
-
76.
Knoops B, Goemaere J, Van der Eecken V, Declercq JP. Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-cys peroxiredoxin. Antioxid Redox Signal. 2011; 15:817–29. https://doi.org/10.1089/ars.2010.3584 [PubMed]
-
77.
Kim B, Kim YS, Ahn HM, Lee HJ, Jung MK, Jeong HY, Choi DK, Lee JH, Lee SR, Kim JM, Lee DS. Peroxiredoxin 5 overexpression enhances tumorigenicity and correlates with poor prognosis in gastric cancer. Int J Oncol. 2017; 51:298–306. https://doi.org/10.3892/ijo.2017.4013 [PubMed]
-
78.
Sevilla F, Camejo D, Ortiz-Espín A, Calderón A, Lázaro JJ, Jiménez A. The thioredoxin/peroxiredoxin/sulfiredoxin system: current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J Exp Bot. 2015; 66:2945–55. https://doi.org/10.1093/jxb/erv146 [PubMed]
-
79.
Veal EA, Underwood ZE, Tomalin LE, Morgan BA, Pillay CS. Hyperoxidation of peroxiredoxins: gain or loss of function? Antioxid Redox Signal. 2018; 28:574–90. https://doi.org/10.1089/ars.2017.7214 [PubMed]
-
80.
Vance TM, Azabdaftari G, Pop EA, Lee SG, Su LJ, Fontham ET, Bensen JT, Steck SE, Arab L, Mohler JL, Chen MH, Koo SI, Chun OK. Thioredoxin 1 in prostate tissue is associated with gleason score, erythrocyte antioxidant enzyme activity, and dietary antioxidants. Prostate Cancer. 2015; 2015:728046. https://doi.org/10.1155/2015/728046 [PubMed]
-
81.
Rodriguez-Garcia A, Hevia D, Mayo JC, Gonzalez-Menendez P, Coppo L, Lu J, Holmgren A, Sainz RM. Thioredoxin 1 modulates apoptosis induced by bioactive compounds in prostate cancer cells. Redox Biol. 2017; 12:634–47. https://doi.org/10.1016/j.redox.2017.03.025 [PubMed]
-
82.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009; 55:611–22. https://doi.org/10.1373/clinchem.2008.112797 [PubMed]
-
83.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001; 25:402–08. https://doi.org/10.1006/meth.2001.1262 [PubMed]
-
84.
Gabriel Kuniyoshi ML, Nunes Da Silva-Gomes R, Cavalcante Souza Vieira J, Casemiro Hessel M, Assunção Mareco E, Dos Santos VB, Carvalho RF, De Magalhães Padilha P, Dal-Pai-Silva M. Proteomic analysis of the fast-twitch muscle of pacu (Piaractus mesopotamicus) after prolonged fasting and compensatory growth. Comp Biochem Physiol Part D Genomics Proteomics. 2019; 30:321–332. https://doi.org/10.1016/j.cbd.2019.04.005 [PubMed]
-
85.
Dionizio AS, Melo CG, Sabino-Arias IT, Ventura TM, Leite AL, Souza SR, Santos EX, Heubel AD, Souza JG, Perles JV, Zanoni JN, Buzalaf MA. Chronic treatment with fluoride affects the jejunum: insights from proteomics and enteric innervation analysis. Sci Rep. 2018; 8:3180. https://doi.org/10.1038/s41598-018-21533-4 [PubMed]
-
86.
da Silva-Gomes RN, Gabriel Kuniyoshi ML, Oliveira da Silva Duran B, Thomazini Zanella BT, Paccielli Freire P, Gutierrez de Paula T, de Almeida Fantinatti BE, Simões Salomão RA, Carvalho RF, Delazari Santos L, Dal-Pai-Silva M. Prolonged fasting followed by refeeding modifies proteome profile and parvalbumin expression in the fast-twitch muscle of pacu (piaractus mesopotamicus). PLoS One. 2019; 14:e0225864. https://doi.org/10.1371/journal.pone.0225864 [PubMed]
-
87.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976; 72:248–54. https://doi.org/10.1006/abio.1976.9999 [PubMed]
-
88.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–97. https://doi.org/10.1093/nar/gkw377 [PubMed]
-
89.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–52. https://doi.org/10.1093/nar/gku1003 [PubMed]
-
90.
Jurmeister S, Ramos-Montoya A, Sandi C, Pértega-Gomes N, Wadhwa K, Lamb AD, Dunning MJ, Attig J, Carroll JS, Fryer LG, Felisbino SL, Neal DE. Identification of potential therapeutic targets in prostate cancer through a cross-species approach. EMBO Mol Med. 2018; 10:e8274. https://doi.org/10.15252/emmm.201708274 [PubMed]
-
91.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017; 45:W98–102. https://doi.org/10.1093/nar/gkx247 [PubMed]
-
92.
Starruß J, de Back W, Brusch L, Deutsch A. Morpheus: a user-friendly modeling environment for multiscale and multicellular systems biology. Bioinformatics. 2014; 30:1331–32. https://doi.org/10.1093/bioinformatics/btt772 [PubMed]
-
93.
Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014; 30:2811–12. https://doi.org/10.1093/bioinformatics/btu393 [PubMed]