Introduction
Traditionally, in most common cancers, including breast cancer (BC), clinicopathological features (e.g., tumor size, lymph node status, TNM stage, histological grade, hormone receptor status, human epidermal growth factor receptor 2 [HER-2] amplification) are used to predict patient outcome [1]. Biomarkers such as tumor-associated macrophages (TAMs), microRNAs, matrix metalloproteinases (MMPs), retinoic acid receptor a (RARA), and Ki-67 are also useful in predicting prognosis of certain cancers (e.g., colon cancer, gastric cancer, acute promyelocytic leukemia) [2–6]. There has been increasing research focus on protein glycosylation and related glycosyltransferases as prognostic biomarkers in various human cancers [7–9].
Glycosylation (attachment of glycans to proteins or other organic molecules) is a common posttranslational modification in all organisms. Aberrant glycosylation is a characteristic phenomenon in carcinogenesis, plays essential roles in specific steps of tumor development [10], and directly promotes tumor progression and metastasis [11–17]. Glycosylation is mediated by enzymatic activities of glycosyltransferases and glycosidases in glycoproteins and/or lipids. Alterations of glycosyltransferase expression are associated with both pro-metastatic and metastasis-suppressing functions [12].
Fucosyltransferase 8 (FUT8) is an α(1,6)-fucosyltransferase responsible for addition of fucose to asparagine-linked N-acetylglucosamine (GlcNAc) moieties, a common feature of N-linked glycan core structures [18]. Aberrant fucosylated glycan structures and associated glycosyltransferases have been observed in development of various cancers [19]; e.g., increased core fucosylation in BC [20], non-small cell lung cancer (NSCLC) [21], ovarian cancer [22], colon cancer [23], prostate cancer [24], and melanoma [25]. A positive feedback mechanism of FUT8-mediated receptor core fucosylation was recently shown to enhance TGF-β signaling and epithelial-mesenchymal transition (EMT), thus promoting BC cell invasion and metastasis; such fucosylation in BC patients is a potential diagnostic/ prognostic biomarker, or therapeutic target [26]. In BC patients, high FUT8 protein expression is correlated with lymphatic metastasis and stage status, whereas reduced FUT8 expression is correlated with disease-free survival and overall survival [20].
FUT8 expression level has been linked to tumor clinical features and outcomes in numerous studies. However, few attempts have been made to systematically evaluate such associations. We performed a systematic review and meta-analysis based on collection of references and Gene Expression Omnibus (GEO) microarray data, to clarify the correlations between FUT8 expression and clinical pathology and patient survival in various common types of cancer.
Results
Characteristics of studies included in the meta-analysis
A total of 366 articles were found through database mining and manual searches (see M&M); these comprised 152 articles directly related to FUT8, and 214 articles that included microarray data and were indirectly related to FUT8. After screening out duplicate titles and abstracts, 27 full-text articles and 78 datasets remained; of these, 20 articles and 43 datasets were excluded on the basis of criteria described in M&M, finally leaving 7 articles and 35 microarray datasets (involving 6124 patients) for inclusion in the meta-analysis. Characteristics and quality scores of the included studies are summarized in Table 1. Among these, the 35 datasets involved 7 types of malignant tumors and descriptions of 29 types of clinicopathological features related to FUT8, and the 7 articles involved 5 types of malignant tumors and described correlations between FUT8 expression and patient survival.
Table 1. Characteristics of studies included in the meta-analysis.
| Tumor source | First author and year | Country | Ethnicity | Number of patients | Sample Type | Method | GEO ID | Cutoff | Survival | Follow-up (Months) | Quality Score | Reference |
| Breast Cancer | Lasham 2012 | New Zealand | Caucasian | 107 | tissue | Microarray | gse36771 | average | NR | NR | 7 | [49] |
| Chin 2006 | America | Caucasian | 130 | tissue | Microarray | gse69031 | average | NR | NR | 7 | [50] |
| Desmedt 2009 | Belgium | Caucasian | 55 | tissue | Microarray | gse16391 | average | NR | NR | 6 | [51] |
| EXPO 2005 | America | Caucasian | 351 | tissue | Microarray | gse2109 | average | NR | NR | 8 | R2 platform |
| Lu 2008 | America | Caucasian | 123 | tissue | Microarray | gse5460 | average | NR | NR | 6 | [52] |
| Concha 2011 | Spain | Caucasian | 66 | tissue | Microarray | gse29431 | average | NR | NR | 7 | R2 platform |
| Iwamoto 2011 | America | Caucasian | 103 | tissue | Microarray | gse22093 | average | NR | NR | 7 | [53] |
| Yue 2016 | China | Asian | 189 | tissue | IHC | | median | OS, DFS | 72 | 7 | [20] |
| Colorectal Cancer | EXPO 2005 | America | Caucasian | 315 | tissue | Microarray | gse2109 | average | NR | NR | 7 | R2 platform |
| Laibe 2012 | France | Caucasian | 130 | tissue | Microarray | gse37892 | average | NR | NR | 7 | [54] |
| Jorissen 2009 | Australia | Caucasian | 290 | tissue | Microarray | gse14333 | average | NR | NR | 8 | [55] |
| Smith 2010 | America | Caucasian | 232 | tissue | Microarray | gse17538 | average | NR | NR | 7 | [56] |
| Watanabe 2006 | Japan | Asian | 84 | tissue | Microarray | gse4554 | average | NR | NR | 5 | [57] |
| Tsukamoto 2011 | Japan | Asian | 148 | tissue | Microarray | gse21510 | average | NR | NR | 7 | [58] |
| Jorissen 2008 | Denmark | Caucasian | 155 | tissue | Microarray | gse13294 | average | NR | NR | 6 | [59] |
| Schlicker 2012 | United Kingdom | Caucasian | 62 | tissue | Microarray | gse35896 | average | NR | NR | 5 | [60] |
| Barras 2017 | Australia | Caucasian | 59 | tissue | Microarray | gse75316 | average | NR | NR | 5 | [61] |
| Ependymoma | Donson 2009 | America | Caucasian | 19 | tissue | Microarray | gse16155 | average | NR | NR | 7 | [62] |
| Johnson 2010 | America | Caucasian | 83 | tissue | Microarray | NR | average | NR | NR | 7 | [63] |
| Hoffman 2014 | America | Caucasian | 65 | tissue | Microarray | gse50385 | average | NR | NR | 7 | [64] |
| Vladoiu 2019 | Germany | Caucasian | 209 | tissue | Microarray | gse64415 | average | NR | NR | 8 | [65] |
| Glioma | Freije 2004 | America | Caucasian | 85 | tissue | Microarray | gse4412 | average | NR | NR | 6 | [66] |
| Gravendeel 2009 | Netherlands | Caucasian | 276 | tissue | Microarray | gse16011 | average | OS | 240 | 7 | [67] |
| Kawaguchi 2013 | Japan | Asian | 50 | tissue | Microarray | gse43378 | average | NR | NR | 6 | [68] |
| Zhang 2014 | America | Caucasian | 21 | tissue | Microarray | gse50774 | average | NR | NR | 6 | [69] |
| Non-Small Cell Cancer | Tarca 2013 | Switzerland | Caucasian | 150 | tissue | Microarray | gse43580 | average | NR | NR | 7 | [70] |
| Muley 2014 | Germany | Caucasian | 100 | tissue | Microarray | gse33532 | average | NR | NR | 8 | R2 platform |
| Honma 2015 | Japan | Asian | 129 | tissue | IHC | | median | OS | 168 | 7 | [71] |
| Chen 2013 | China | Asian | 140 | tissue | IHC | | median | OS, DFS | 120 | 7 | [21] |
| Wu 2019 | China | Asian | 135 | tissue | IHC | | median | OS, DFS | 60 | 7 | [72] |
| Park 2020 | Korea | Asian | 217 | tissue | Microarray | gse31210 | median | DFS | 120 | 8 | [73] |
| Medulloblastoma | Robinson 2012 | America | Caucasian | 76 | tissue | Microarray | gse37418 | average | NR | NR | 6 | [74] |
| Northcott 2017 | Germany | Caucasian | 223 | tissue | Microarray | NR | average | NR | NR | 7 | [75] |
| Kool 2008 | Netherlands | Caucasian | 62 | tissue | Microarray | gse10327 | average | NR | NR | 7 | [76] |
| Delattre 2012 | NR | NR | 57 | tissue | Microarray | NR | average | NR | NR | 6 | R2 platform |
| Neuroblastoma | Delattre 2009 | France | Caucasian | 34 | tissue | Microarray | gse14880 | average | NR | NR | 5 | R2 platform |
| Ohtaki 2010 | Japan | Asian | 51 | tissue | Microarray | gse16237 | average | NR | NR | 6 | [77] |
| Lastowska 2007 | United Kingdom | Caucasian | 30 | tissue | Microarray | gse13136 | average | NR | NR | 5 | [78] |
| Molenaar 2012 | Netherlands | Caucasian | 88 | tissue | Microarray | gse16476 | average | NR | NR | 7 | [79] |
| Pancreatic Ductal Adenocarcinoma | Tada 2019 | Japan | Asian | 62 | tissue | IHC | | median | RFS | 120 | 7 | [80] |
| Diffuse Large B Cell Lymphoma | Xiao 2008 | America | Caucasian | 420 | tissue | Microarray | gse10846 | average | OS | 240 | 8 | R2 platform |
| Gastric Cancer | Tan 2018 | Switzerland | Caucasian | 192 | tissue | Microarray | gse15459 | average | OS | 60 | 7 | R2 platform |
| OS, overall survival; DFS, disease-free survival; RFS, relapse-free survival; IHC, immunohistochemistry; NR, not reported. |
Association of FUT8 expression with clinicopathological features of various types of malignant tumors
Seven malignant tumors (ependymoma, glioma, BC, colorectal cancer (CRC), medulloblastoma (MBL), neuroblastoma (NB), NSCLC) were included in meta-analyses for clinicopathological features. Pooled results are presented in Supplementary Table 1. For BC, high FUT8 expression level was related to positive PR and positive ER status (odds ratio [OR]= 3.34, 95% confidence interval [CI]: 1.60-6.96, p= 0.001 and odds ratio [OR]= 7.42, 95% confidence interval [CI]: 2.94-18.7, p< 0.0001). FUT8 expression level was also correlated with tumor histological grade (OR= 2.55, 95% CI: 1.10-5.95, p= 0.03) (Figure 1A). For CRC, elevated FUT8 expression level was associated with TNM stage I-III (OR= 1.79, 95% CI: 1.09–2.95, p= 0.02), and was also related to microsatellite instability (MSI) (OR= 4.37, 95% CI: 2.65-7.20, p< 0.00001) and female gender (OR= 0.65, 95% CI: 0.50-0.83, p= 0.0007) (Figure 1B). For ependymoma, high FUT8 expression level was related to patient age (≤10 years) (OR= 3.69, 95% CI: 2.28-5.99, p< 0.00001) and tumor recurrence status (OR= 2.29, 95% CI: 1.07-4.93, p= 0.03) (Supplementary Figure 1A). For glioma, increased FUT8 level was associated with glioblastoma multiforme (GBM) (OR= 1.69, 95% CI: 1.07–2.66, p= 0.02), and FUT8 level was inversely correlated with patient age (≤40 years) (OR= 0.58, 95% CI: 0.35-0.95, p= 0.03) (Supplementary Figure 1B). For MBL, high FUT8 expression level was related to Wingless (WNT) (OR= 4.72, 95% CI: 1.99-11.22, p= 0.0004) and Sonic Hedgehog (SHH) molecular subgroups (OR= 12.11, 95% CI: 6.44-22.79, p< 0.00001). FUT8 level was inversely correlated with metastasis status (OR= 0.25, 95% CI: 0.10-0.60, p= 0.002) and male gender (OR= 0.60, 95% CI: 0.39-0.92, p= 0.02) (Figure 2A). For NSCLC, FUT8 expression level was inversely correlated with N0 status of TNM stage (OR= 0.58, 95% CI: 0.38-0.88, p= 0.01) and male gender (OR= 0.34, 95% CI: 0.18-0.68, p= 0.002) (Figure 2B). Clinicopathological features of malignant tumors related to FUT8 are summarized in Table 2. High FUT8 expression level in NB showed no association with any clinicopathological feature, including INSS stage, gender, or mycn amplified status (Supplementary Figure 2).
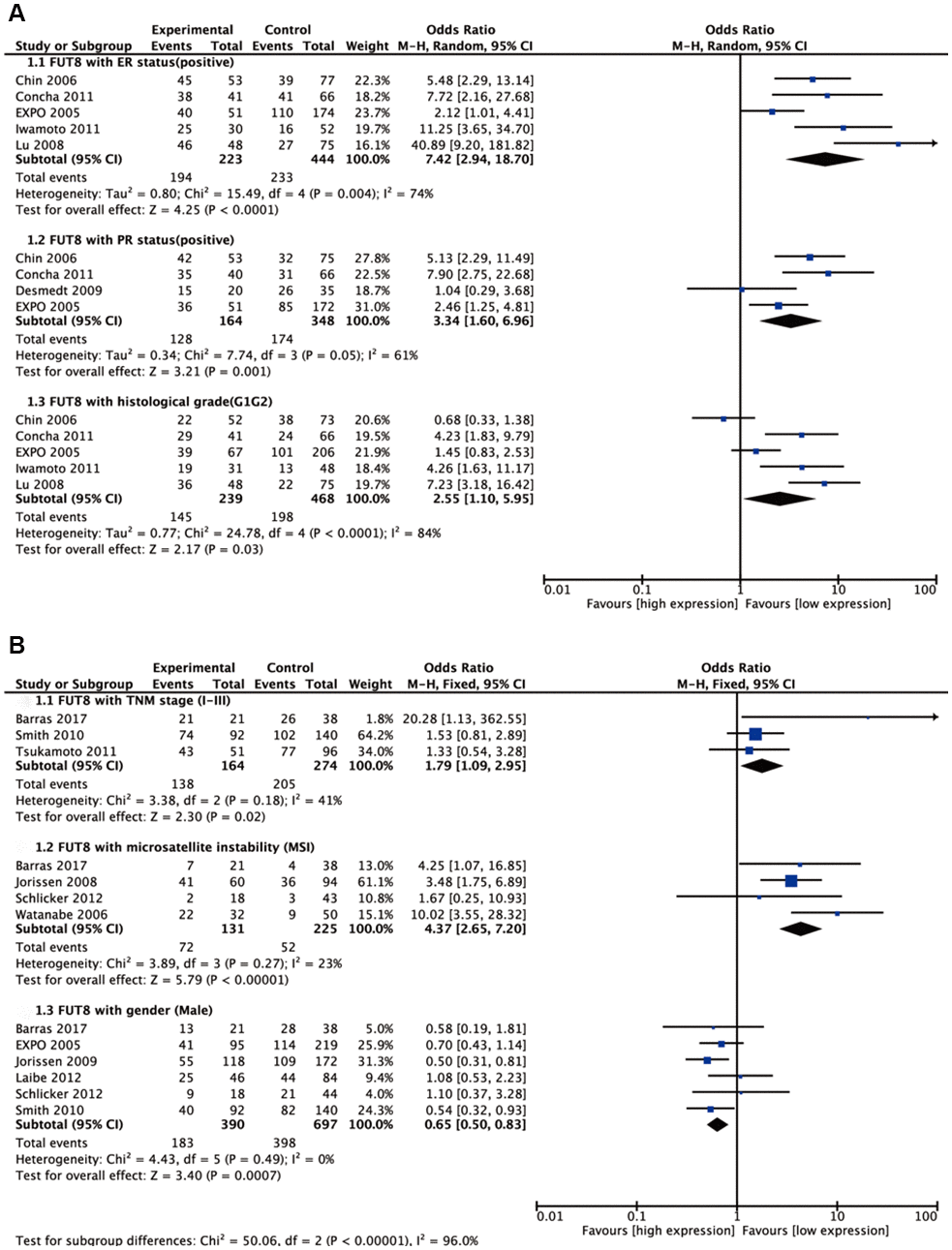
Figure 1. Forest plots of the significant associations between FUT8 expression and clinical features in two tumor types. (A) breast cancer; (B) colorectal cancer.
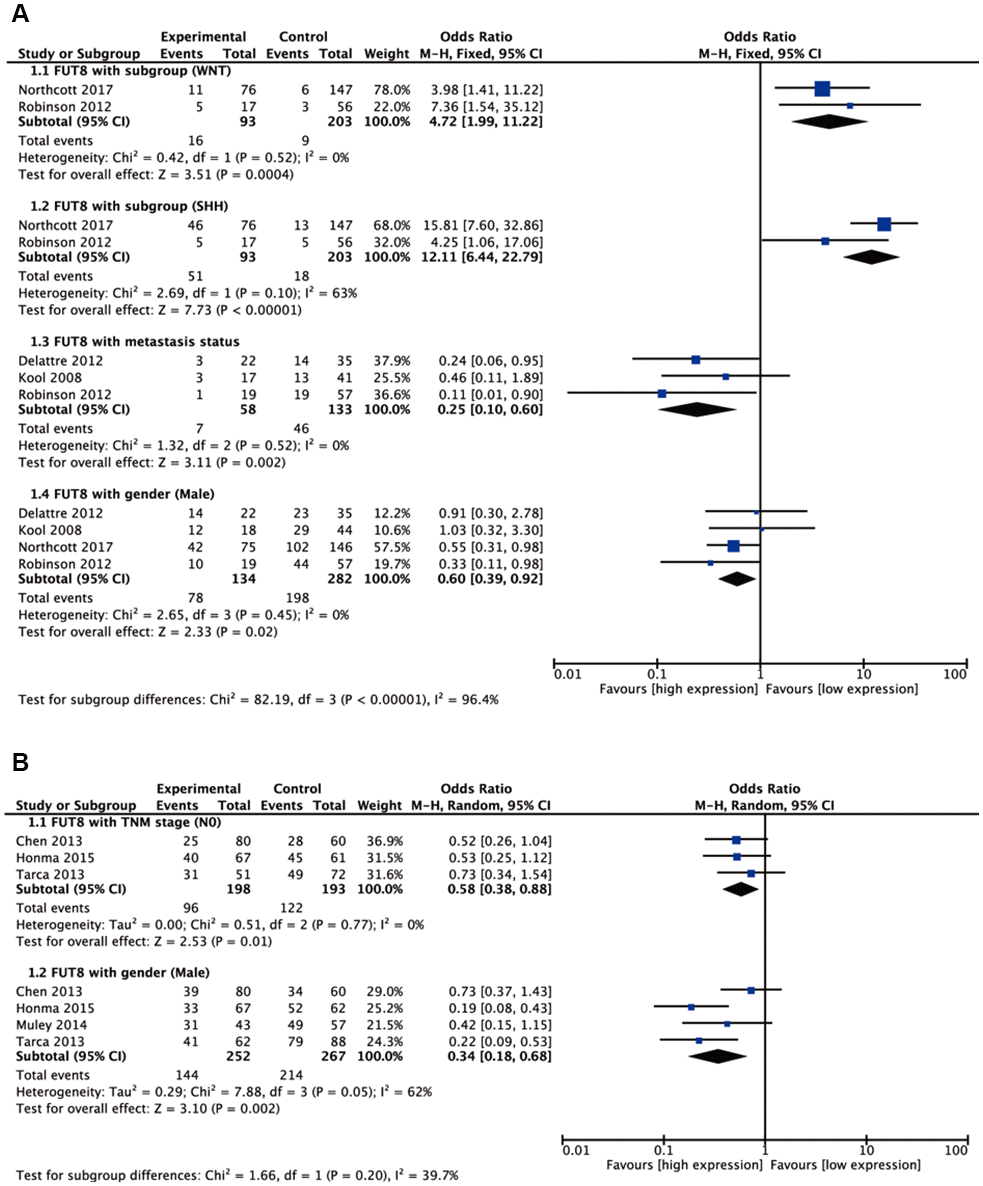
Figure 2. Forest plots of significant associations between FUT8 expression and clinical features in two tumor types. (A) medulloblastoma; (B) non-small cell lung cancer.
Table 2. Clinicopathological features related to enhanced FUT8 expression level in various malignant tumors.
| Tumor source | Clinicopathological features | Reference |
| Breast cancer | ER status positive, PR status positive, histological grade (G1G2) histological grade (G1) | [50], [51], [52], [53], R2 platform |
| Colorectal cancer | Duke stage A, Duke stage AB, location (Left), gender (male) microsatellite instability (MSI) | [54], [55], [56], [57], [58], [59], [60], [61], R2 platform |
| Glioma | Age (≤40 years), glioblastoma multiforme (GBM) | [66], [67], [68], [69] |
| Medulloblastoma | Molecular subgroups (G3), molecular subgroups (G4), molecular subgroups (SHH), molecular subgroups (WNT) Gender (male), metastasis | [74], [75], [76], R2 platform |
| Non-small cell lung cancer | Gender (male), TNM stage N0, TNM stage M1 | [21], [70], [71], R2 platform |
| Ependymoma | Age (≤10 years), recurrence | [63], [64], [65] |
| ER, estrogen receptor; PR, progesterone receptor. |
Prognostic value of FUT8 expression in survival of various types of malignant tumors
Five types of malignant tumors (NSCLC, glioma, diffuse large B cell lymphoma (DLBCL), gastric cancer (GC), BC) were included in meta-analyses for survival. For NSCLC, elevated FUT8 expression level was associated with shorter disease-free survival (DFS) (hazard ratio [HR]= 2.32, 95% CI: 1.65-3.27, p< 0.00001), and with lower overall survival (OS) (HR= 2.24, 95% CI: 1.62-3.08, p< 0.00001) (Figure 3).
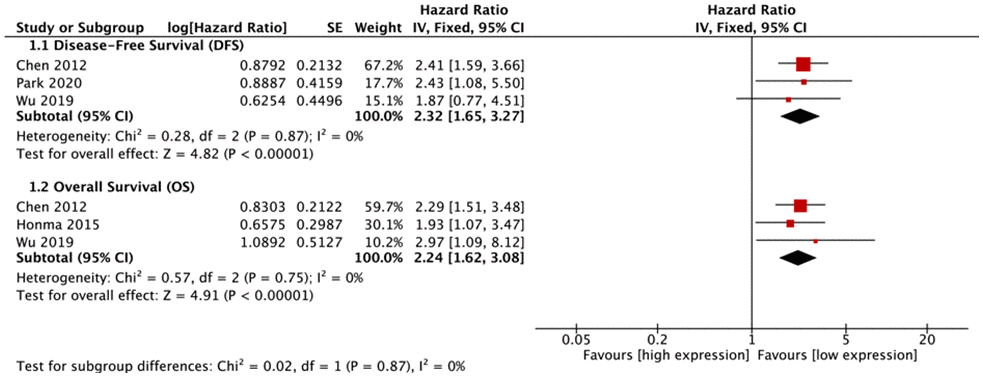
Figure 3. Forest plots of associations between FUT8 expression and non-small cell lung cancer overall survival.
FUT8 expression levels in the other 4 tumor types (glioma, DLBCL, GC, BC) were also correlated with OS (Figure 4). Among these, high FUT8 expression was associated with shorter OS in glioma (RR= 1.49, 95% CI: 1.13-1.96), DLBCL (RR= 1.76, 95% CI: 1.30-2.38), and BC (RR= 2.49, 95% CI: 1.15-5.38), but with longer OS in GC (RR= 0.60, 95% CI: 0.40-0.91). For glioma, upregulated FUT8 expression was associated with shorter OS in female patients (RR= 1.99, 95% CI: 1.15-3.44). For DLBCL, high FUT8 expression was associated with shorter OS in Ann Arbor stages I-III (RR= 1.87, 95% CI: 1.24-2.81) and in patients aged >50 years (RR= 1.42, 95% CI: 1.02-1.98). For GC, high FUT8 expression was associated with better OS in intestinal-type Lauren classification (RR= 0.50, 95% CI: 0.28-0.90), males (RR= 0.59, 95% CI: 0.35-0.99), and TNM stage I-III patients (RR= 0.53, 95% CI: 0.31-0.90). Pooled survival results are presented in Supplementary Table 2.
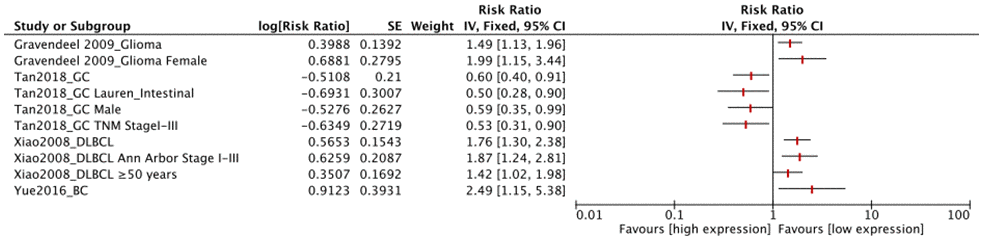
Figure 4. Forest plots of associations between FUT8 expression and tumor overall survival in single clinical features studied.
Discussion
FUT8 is clearly involved in tumor initiation and progression, and in various biological behaviors of cancer, including cell proliferation, apoptosis, migration, and metastasis [27]. Prognostic values of glycosylation or fucosylated antigens in many types of cancer have been documented in several previous reviews and meta-analyses [14, 15, 28–30]. However, very little meta-analysis of FUT8 has been performed, because of lack of sufficient studies and references. Many reports are based on use of microarrays to study cancer genomes; accordingly, we performed a systematic review after collecting GEO microarray data, and data from a large number of studies, focused on the prognostic value of FUT8 in cancer. This review / meta-analysis is, to our knowledge, the first to comprehensively clarify the association of FUT8 expression with specific clinicopathological features and survival data for various types of cancer.
Our analyses demonstrated that FUT8 expression levels were most often associated with tumor stage (n=4: BC, CRC, MBL, NSCLC), molecular classification (n=2: BC, MBL), age (n=2: glioma, ependymoma), and gender (n=2: MBL, NSCLC), but less associated with histological grade (n=1: BC) and pathological typing (n=1: glioma). FUT8 expression showed no association with disease induction, location, or family history for any of the above cancer types. These findings suggest that FUT8 plays an intrinsic role mainly in tumor development, and is therefore a potentially important biomarker for malignant tumors. Previous studies have focused mainly on the relationship between FUT8 expression and OS for survival. The majority of such studies (n=5: NSCLC, BC, DLBCL, GC, glioma) found significant correlations of FUT8 expression with OS of tumor, and also associations between survival and clinicopathological features, particularly tumor stage. A few studies focused on associations of FUT8 expression with other types of survival (DFS, RFS). For the research on pathways related to FUT8 and tumor survival, many references indicate that FUT8 activates several related signaling pathways, including Ras/MAPKK signaling, c-Met signaling, Akt/mTOR signaling and Wnt/β-catenin signaling, which ultimately lead to hepatocellular carcinoma or colorectal cancer affects the overall survival of patients [31–33]. Höti et al demonstrated that overexpression of FUT8 resulted in upregulation of epidermal growth factor receptor (EGFR) and corresponding downstream signaling, leading to increased prostate cancer cells survival [34]. In short, findings of these studies, taken together, demonstrate the strong prognostic value of FUT8 expression in malignant tumors.
NSCLC is the tumor type most frequently studied in regard to FUT8 expression status. High FUT8 expression was inversely correlated with N0 status of TNM stage and with male gender. High FUT8 expression was associated with shorter DFS, and with lower OS. Thus, FUT8 should be a useful survival prognostic predictor for female NSCLC patients with high FUT8 expression. Our analyses revealed an association of high FUT8 expression level with positive ER and PR status, and with shorter OS, in BC patients. FUT8 expression is therefore presumably enhanced in luminal A and B BC patients, and may be a useful survival prognostic predictor in such patients. A 2019 study indicated that FUT8 is highly expressed in ER+ BC patients, is associated with metastasis, and is a potential therapeutic target in these patients [35].
Microsatellite instability (MSI) is a molecular "fingerprint" of defective mismatch repair systems, and methods to detect MSI are well established and routinely incorporated into clinical practice. Prognosis for MSI tumors is better than that for microsatellite stable CRC [36]. In regard to CRC, we found that high FUT8 expression was associated with MSI, female gender, and TNM stages I-III. E-cadherin, a Ca2+-dependent cell adhesion molecule, was found to be significantly enhanced in dense culture of FUT8-transfected colorectal adenocarcinoma cells, resulting in increased cell-cell adhesion [23]. In addition, E-cadherin truncation was significantly higher in low MSI as compared with high MSI tumors [37]. The inferred mechanism is that the low expression of FUT8 in TNM stages IV or microsatellite stable colorectal cancer may cause E-cadherin to decrease or be truncated, resulting in decreased tumor cell-cell adhesion and increased metastasis.
Medulloblastoma, a common type of malignant brain cancer that accounts for 8-10% of childhood brain tumors [38], comprises four molecular subgroups (WNT, SHH, group 3, group 4) that have differing prognoses [39]. In general, subgroup WNT has very good prognosis and rare metastasis; subgroup SHH has good prognosis in infants and intermediate prognosis and uncommon metastasis in older children; subgroup G4 has intermediate prognosis and frequent metastasis; subgroup G3 has poor prognosis and very frequent metastasis [40–43]. Our analysis revealed association of high FUT8 expression with subgroups WNT and SHH, and inverse correlation with metastasis and male gender. High FUT8 expression in medulloblastoma therefore appears to be a positive prognostic indicator, particularly for female patients. In regard to NB, we did not find any notable relationship of high FUT8 expression with clinicopathological features. On the other hand, another group reported aberrant expression of N-glycans and short O-glycans in NB cells, and regulation of their expression levels by associated glycosyltransferases [44]. GnT-V expression in NB patients was correlated with favorable prognosis and treatment outcome [45]. GALNT9 was expressed in neuroblasts derived from primary tumor [46] but not in those derived from metastatic bone marrow, and may be a useful prognostic marker for positive clinical outcome in NB patients [47].
Overall, our results are comprehensive and seemingly reliable in view of the high quality of included articles and microarray data. On the other hand, there are inherent limitations in our analysis. (i) Heterogeneity is present among studies of a given tumor type, and is difficult to address because of methodological differences (e.g., sample selection, detection method, determination of cutoff value, statistical analysis). (ii) Nearly all our included studies report a statistically significant result. Although a Begg’s funnel plot indicates absence of publication bias (Supplementary Figure 3), our experience suggests that selective reporting bias is common in the literature regarding FUT8 and tumor prognosis. (iii) Roughly half of our included studies had small sample size (<100), which is often associated with inflated estimates of effect size and with high heterogeneity.
Results of our analyses – with due consideration of the above caveats – highlight the prognostic value of FUT8 expression in a variety of malignant tumors, and the important biological function of FUT8 in tumor progression. FUT8 may exert its effects in such tumors by regulating functional protein core fucosylations involved in tumor development and metastasis (e.g., L1CAM, P53, TGF-β, EGFR) [25, 34, 48]. Thus, dysregulation of FUT8 would have effects on tumor development, and consequently on clinical prognosis of patients. The molecular mechanisms remain to be clarified, and are being addressed in ongoing studies.
In conclusion, our systematic review and meta-analysis confirmed the association of FUT8 with clinicopathological features and patient survival rates for certain malignant tumor types. FUT8 expression is a significant predictor of prognosis for these tumors. The relative weight of FUT8 correlations with particular clinical features is currently difficult to evaluate because of the presence of many uncontrollable factors. Reliable verification of prognostic value of FUT8 in these tumor types will require a large-scale study using standardized methods of detection, analysis, and reporting.
Materials and Methods
We performed this review based on PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) criteria.
Search strategy
The PubMed, Embase, Web of Science, Springer, Chinese National Knowledge Infrastructure (CNKI), and Wan Fang databases were searched systematically to identify and retrieve all pertinent publications through June 24, 2020. Keywords and search terms used were: FUT8 or Fut8 or Fucosyltransferase 8 or α1-6 Fucosyltransferase or Core fucosylation or Fucosyltransferase; Malignant tumor or Cancer or Carcinoma or Neoplasm; and clinical or clinicopathological or clinicopathology or prognosis predictor or survival or Odds Ratio (OR) or Hazard Ratio (HR). References in retrieved articles were screened manually. Languages of retrieved articles were restricted to English and Chinese.
Data extraction and analysis of GEO datasets
Our first step was to read and screen articles related to tumor clinicopathological features and prognosis, and search for microarray datasets through the references. In this way, 96 datasets from 214 articles were extracted for preliminary screening. The second step was to re-screen the 96 datasets in regard to presence of FUT8 data, an appropriate clinical pathology track, and sufficient number of samples. In this way, 35 malignant tumor microarray datasets were selected and downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds) and R2 platform (http://r2.amc.nl). These datasets included a total of 4918 samples, from 7 types of malignant tumor: breast cancer (BC), colorectal cancer (CRC), ependymoma, glioma, non-small cell lung cancer (NSCLC), medulloblastoma (MBL), and neuroblastoma (NB). The datasets all used the Affymetrix Human Genome U133a or U133 Plus 2.0 expression arrays to detect expression value signals. Data were normalized using Microarray Suite (MAS) V. 5.0. In many cases, more than one probeset has been reported for FUT8 gene. We selected a probeset with highest average presence signal (APS) by default. mRNA expression of FUT8 was assigned to "low" or "high" category based on average expression value of each dataset.
Inclusion and exclusion criteria
Studies were included if they met the following criteria: (1) original study focused on human subjects; (2) presented FUT8 expression data and malignant tumor clinicopathological features and/or survival data; (3) reported an OR or HR with 95% confidence interval (CI), or sufficient data were presented so that we could calculate them; (4) full text was available. Exclusion criteria were as follows: (1) study lacked key information (e.g., clinical parameters, survival curves), or usable data; (2) APS of probeset was too low, or too many data were lost; (3) HRs applied to a combination of multiple FUTs; (4) article was a review, letter, single case report, or conference abstracts. In cases of multiple articles from the same group, reporting overlapping data, only the most complete one was included. The article selection process is shown in flow diagram form in Figure 5.
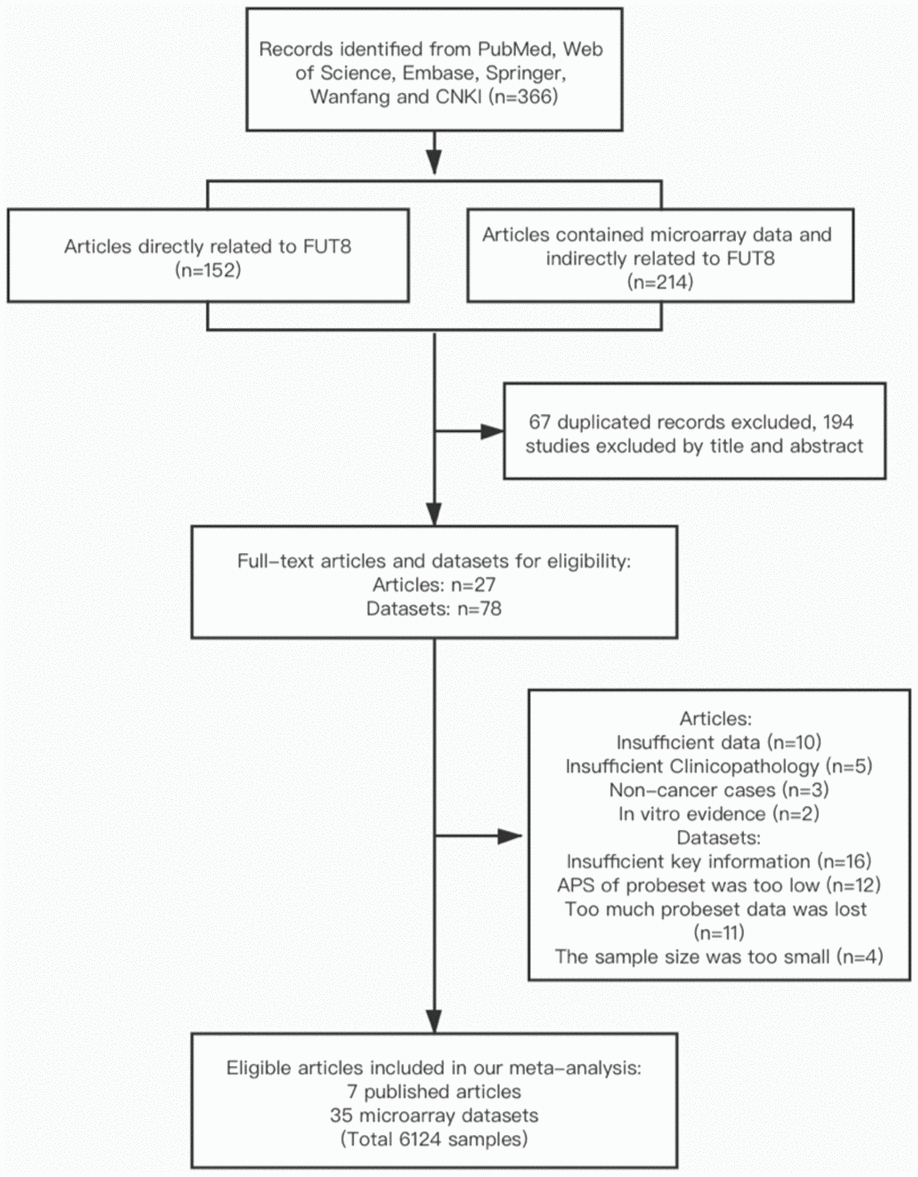
Figure 5. Flow diagram of article selection process.
Quality assessment and data extraction
Two authors (MXM and GXH) reviewed potentially eligible articles independently. Quality of each study was assessed using the Newcastle-Ottawa Scale. The following information was extracted from each included study:
(1) basic information: first author’s or uploaded dataset author’s name, publication year, country of origin, names of malignant tumors, sample size, FUT8 expression levels, detection methods, sample type, outcome measurements, follow-up duration, cutoff value, survival analysis method; (2) p values for correlation between FUT8 expression and clinicopathological features of malignant tumors, and original data used for calculation of ORs and their 95% CIs; (3) HRs and their 95% CIs for survival analysis. If HRs were not directly accessible in the text, Kaplan-Meier survival curves were read using Engauge Digitizer (V. 4.1) to obtain data. Different datasets for a particular malignant tumor were considered as separate studies, and respective HRs were extracted. In cases of possible discrepancy, a consensus was reached by discussion among all authors.
Statistical analysis
ORs and their 95% CIs were used to estimate associations of FUT8 with clinical features of malignant tumors. For purposes of comparison, patients were divided into paired categories (e.g., male vs. female; TNM stages I, II and III vs. IV; ER/PR status positive vs. negative). For survival rates, HRs with corresponding 95% CIs were used. All ORs and HRs were calculated for high FUT8 expression. When a given FUT8 was investigated in two or more different studies, a meta-analysis was performed to combine the effect size. Z test was used to determine significance of ORs or HRs. Heterogeneity between studies was tested using Q statistic and I2 test. When I2 value was >50% (indicating significant heterogeneity) a random-effects model was used. For I2 value ≤50%, a fixed-effects model was used. Statistical analyses were performed with software program Review Manager V. 5.3 (Cochrane Collaboration; London, UK). Differences with p< 0.05 were considered statistically significant.
MXM and XL conceived and designed the study. MXM and GXH searched and reviewed literature. MXM, GXH, ZYZ and YW contributed to data collection, analysis, interpretation and prepared tables and figures. MXM drafted the manuscript. XL and FG revised the manuscript. All authors approved the final manuscript.
The authors declare no conflicts of interest.
This study was supported by the Youth Science Fund of the Fifth People’s Hospital of Qinghai Province (No. 2017-Y-01). The present study was supported by the National Natural Science Foundation of China (No. 81770123, 32071274), and Hundred-Talent Program of Shaanxi Province, and Youth Innovation Team of Shaanxi Universities.