Dynamic regulation in multiple signaling pathways
In the study of Youngman MJ et al. [14], a decline in PMK-1 p38 mitogen-activated protein kinase (MAPK) pathway, one of conserved pathways involved in pathogen defense, was observed by analyzing gene expression levels using unpaired t test in synchronized populations of N2 worms during mid-to-late adulthood (at Day 6 of adulthood and Day 15 of adulthood). It was consistent with our result on MAPK signaling pathway, which was identified as significantly down-regulated from D6 to D15 of adulthood (highlighted in red color in Additional file 1). Surprisingly, MAPK signaling pathway was shown to be significantly up-regulated during young-to-mid adulthood (at the late larval L4 stage and Day 6 of adulthood) in our study, which was not reported by Youngman MJ et al. study [14]. Furthermore, MAPK signaling pathway was identified as truly aging-dependent pathways based on the hierarchical clustering of both samples and involved genes (Figure 2A for the data from L4 to D6 and Fig2B from D6 to D15). Obviously, using the expression data of involved genes in MAPK signaling pathway, six samples were clustered into two groups with samples at the same stage (L4-1, L4-2 and L4-3 in group of L4; D6-1, D6-1 and D6-3 in group of D6; D15-1, D15-1 and D15-3 in group of D15). Actually, MAP kinase (MAPK)/Receptor Tyrosine Kinase (RTK)/RasGTPase signaling pathways were used repeatedly during metazoan development, which were reported to be required for larval viability and for many different developmental processes, including induction of vulval, uterine, spicule, P12 and excretory duct cell fates, control of sex myoblast migration and axon guidance, and promotion of germline meiosis [15]. The core components or regulators of C. elegans RTK/Ras/MAPK signaling pathway were also identified as the aging-dependent targets [16-18], such as LET-23, LIN-1 and LIN-45 (Figure 2 and Additional file 2). Among these candidate targets, four interesting genes, including ced-2, lin-45, ZK856.8 and F59D6.7, were significantly up-regulated from L4 to D6 and down-regulated from D6 to D15, showing dynamic expression patterns. Their significances of gene expression changes between two adjacent stages were shown in Table 2. In C. elegans, the ced-2 gene encodes a Src homology (SH) 2 and 3-containing adaptor protein, homologous to human CrkII, which is required for phagocytosis during programmed cell death and for migration of the distal tip cells of the somatic gonad. It was previously reported that ced-2 mutants may exhibit persistant corpses indicating defects in phagocytosis and distal tip cell migration defects in the gonadal arms [19]. The lin-45 gene encodes an ortholog of the vertebrate protein RAF which is required for larval viability, fertility and the induction of vulval cell fates [18]. Based on RNAi studies of three genes constituting the ERK cascade (lin-45/RAF1, mek-2/MEK1/2, andmpk-1/ERK1/2) as well as lip-1 encoding a MAPK phosphatase that inactivates MPK-1, a novel ERK-MAPK-mediated signaling pathway has been identified, which promotes longevity through two candidate transcription factors, SKN-1 and DAF-16 [20]. Additionally, the genes of ZK856.8 and F59D6.7 would be two important targets for further study of aging in MAPK signaling pathway.
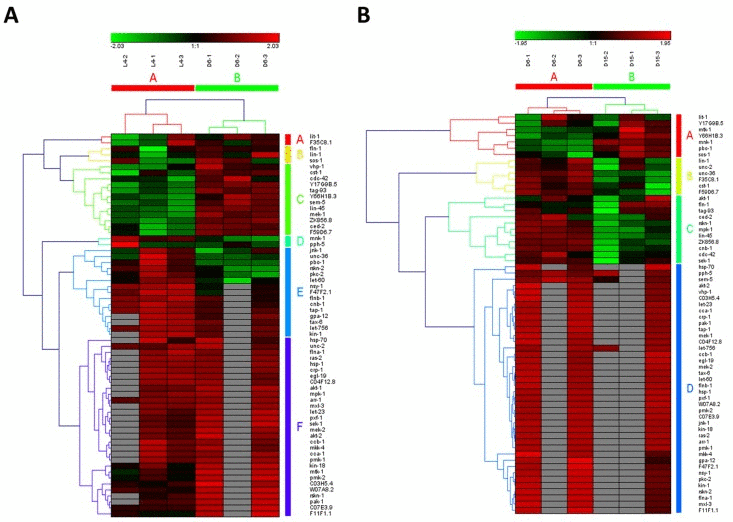
Figure 2. The heat map and hierarchical clustering in MAPK signaling pathway during aging. (A) It showed the heat map and hierarchical clustering in MAPK signaling pathway from the stage of L4 to D6. The samples (column) were clustered into two groups, three replicates in the stage of L4 (L4-1, L4-2 and L4-3) were clustered together and three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together. There were 64 involved genes in MAPK signaling pathway from L4 to D6, which were clustered into 6 groups (the group from A to F).(B) It showed the heat map and hierarchical clustering in MAPK signaling pathway from the stage of D6 to D15. The samples were clustered into two groups, three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together and three replicates at day 15 (D15-1, D15-2 and D15-3) were clustered together. There were also 64 involved genes in MAPK signaling pathway from D6 to D15, which were clustered into 4 groups (the group from A to D). Red is for up-regulated and green is for down-regulated.
Table 2. The significantly associated genes with dynamic regulation pattern in signaling pathways during aging
| Genes | Related pathways | p1 (L4 to D6) | p2 (D6 to D15) |
|---|
| ced-2 | MAPK signaling pathway | 1.70E-02 | 3.62E-04 |
| lin-45 | MAPK signaling pathway mTOR signaling pathway ErbB signaling pathway | 1.34E-03 | 1.87E-03 |
| ZK856.8 | MAPK signaling pathway | 1.03E-03 | 7.11E-03 |
| F59D6.7 | MAPK signaling pathway | 1.41E-02 | 2.17E-02 |
| pdk-1 | mTOR signaling pathway | 4.58E-04 | 1.19E-02 |
| cyd-1 | Wnt signaling pathway | 4.50E-03 | 1.58E-03 |
| sma-4 | Wnt signaling pathway TGF-beta signaling pathway | 9.60E-03 | 5.51E-03 |
| skr-7 | Wnt signaling pathway TGF-beta signaling pathway | 3.76E-06 | 2.05E-02 |
| skr-8 | TGF-beta signaling pathway | 3.12E-05 | 4.59E-02 |
| skr-9 | Wnt signaling pathway TGF-beta signaling pathway | 1.03E-04 | 3.57E-02 |
| skr-13 | Wnt signaling pathway TGF-beta signaling pathway | 1.34E-05 | 2.27E-03 |
| skr-14 | Wnt signaling pathway TGF-beta signaling pathway | 5.99E-05 | 6.63E-03 |
The mammalian target of rapamycin (mTOR), also known as mechanistic target of rapamycin has been identified as a key modulator of ageing and age-related disease. The fact that inhibition of mTOR signaling pathway may extend lifespan in model organisms and confer protection against a growing list of age-related pathologies has been clearly approved [21]. In our study, this pathway was also identified as one of truly aging-dependent pathways with dynamic regulation, which was significantly up-regulated from L4 to D6 and down-regulated from D6 to D15 (Table 1). Like MAPK signaling pathway, based on the expression of candidate genes in mTOR signaling pathway, the samples at the same time point can be clustered together (Figure 3A for the data from L4 to D6 and Figure 3B from D6 to D15). There were 24 genes involved in mTOR signaling pathway both in the period of young-to-mid and mid-to- late adulthood, which were both clustered into 2 groups (the group of A and B in Figure 3A & 3B). Among these genes appearing in both the group of B in Figure 3A and the group A in Figure 3B, besides lin-45 mentioned above, the gene of pdk-1 was identified with dynamic expression pattern, which was significantly up-regulated from L4 to D6 (p=4.58E-04) and down-regulated from D6 to D15 (p=1.19E-02) (Table 2).In C. elegans, pdk-1 encodes the 3-phosphoinositide-dependent kinase 1 ortholog (PDK-1), which is a component of the DAF-2/insulin receptor-like signaling pathway and accordingly, functions to regulate such processes as dauer larvae formation as well as longevity [21]. The increased lifespan in daf-2 insulin/IGF-1 receptor mutants is accompanied by up-regulation of the MDL-1 Mad basic helix-loop-helix leucine zipper transcriptionfactor, which is an inhibitor of cell proliferation and growth that slows progression of an age-related pathology in C. elegans [22]. The previous studies have illustrated that mTOR signaling pathway plays the central role, whereas other pathways act on it by activating or antagonizing either at upstream or downstream [23-26].
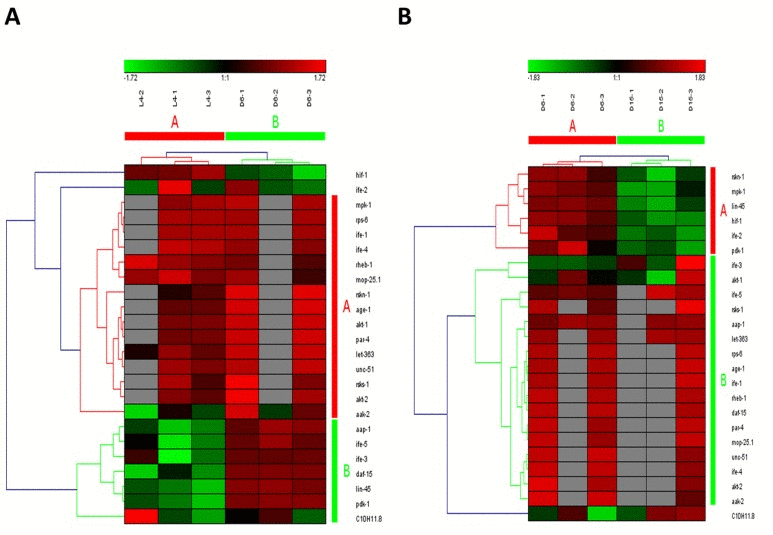
Figure 3. The heat map and hierarchical clustering in mTOR signaling pathway during aging. (A) It showed the heat map and hierarchical clustering in mTOR signaling pathway from the stage of L4 to D6. The samples (column) were clustered into two groups, three replicates in the stage of L4 (L4-1, L4-2 and L4-3) were clustered together and three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together. There were 24 involved genes (row) in mTOR signaling pathway from L4 to D6, which were clustered into 2 groups (the group of A and B). (B) It showed the heat map and hierarchical clustering in mTOR signaling pathway from the stage of D6 to D15. The samples were clustered into two groups, three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together and three replicates at day 15 (D15-1, D15-2 and D15-3) were clustered together. There were also 24 involved genes in mTOR signaling pathway from D6 to D15, which were clustered into 2 groups (the group of A and B).Red is for up-regulated and green is for down-regulated.
Wnt signaling pathways have extremely diverse functions in animals, including the control of gene expression, cell behavior, cell adhesion, and cell polarity [19]. During aging in C. elegans, Wnt signaling pathway was identified as another truly aging-dependent pathway with dynamic regulation, which was significantly up-regulated from L4 to D6 and down-regulated from D6 to D15. There were 62 involved genes in Wnt signaling pathway, which were clustered into 4 groups from L4 to D6 (the group from A to D in Figure 4A) and clustered into 3 groups (the group from A to C in Figure 4B). Among these genes appearing in both the group of A in Figure 4A and the group C in Figure 4B, there were 8 dynamically regulated genes containing sma-4, cyd-1, skr-7,9,13,14,F59D6.7 and ZK856.8, which were significantly up-regulated from L4 to D6 and down- regulated from D6 to D15(Table 2).The gene of sma-4 encodes a Smad protein and a homolog of human DPC4, the protein of SMA-4 is similar to members of the vertebrate protein family of Dwarfins. During development, sma-4 functions as part of a DBL-1/SMA-6 TGF-beta-related signaling pathway that controls body size and male tail sensory ray and spicule formation as well as regulates reproductive aging via oocyte and germline quality maintenance [27]. Indeed, TGF-beta signaling pathway was one of our candidate aging-dependent pathways (Figure 5). The Smad-mediated TGF-beta signaling pathway controls numerous cellular responses from cell proliferation, differentiation and extracellular matrix remodelling to embryonic development in species ranging from worms to mammals [20]. It has been shown that TGF-beta superfamily ligands can regulate cellular or physiological processes through non-canonical pathways by the activation of other signaling molecules, e.g. Akt, MAPK, mTOR, and Src independent of Smad proteins [28]. In this study, Wnt signaling pathway would be seen as one of Smad-dependent pathways in TGF-beta family signaling based on the common regulator of SMA-4 (Table 2).
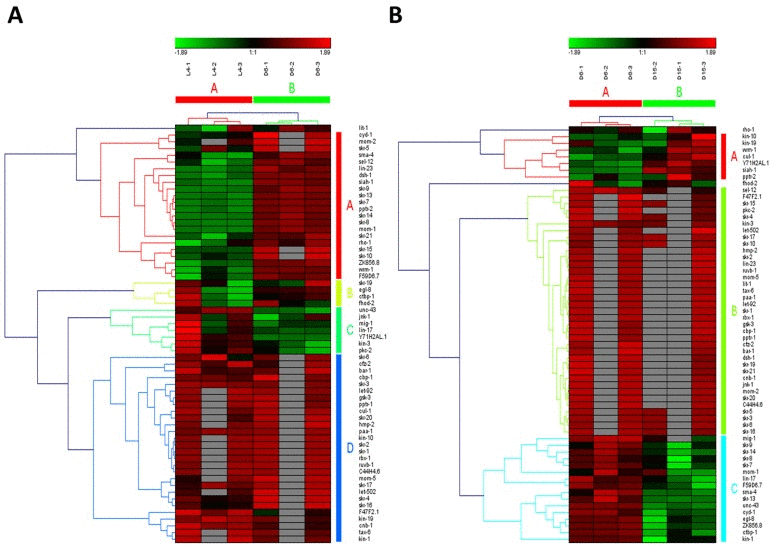
Figure 4. The heat map and hierarchical clustering in Wnt signaling pathway during aging. (A) It showed the heat map and hierarchical clustering in Wnt signaling pathway from the stage of L4 to D6. The samples (column) were clustered into two groups, three replicates in the stage of L4 (L4-1, L4-2 and L4-3) were clustered together and three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together. There were 62 involved genes (row) in Wnt signaling pathway from L4 to D6, which were clustered into 4 groups (the group from A to D). (B) It showed the heat map and hierarchical clustering in Wnt signaling pathway from the stage of D6 to D15. The samples were clustered into two groups, three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together and three replicates at day 15 (D15-1, D15-2 and D15-3) were clustered together. There were also 62 involved genes in Wnt signaling pathway from D6 to D15, which were clustered into 3 groups (the group from A to C). Red is for up-regulated and green is for down-regulated.
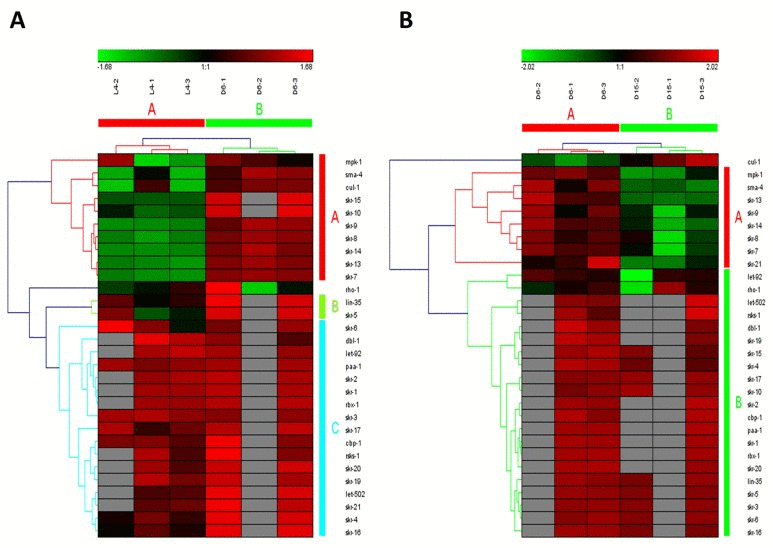
Figure 5. The heat map and hierarchical clustering in TGF-beta signaling pathway during aging. (A) It showed the heat map and hierarchical clustering in TGF-beta signaling pathway from the stage of L4 to D6. The samples (column) were clustered into two groups, three replicates in the stage of L4 (L4-1, L4-2 and L4-3) were clustered together and three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together. There were 30 involved genes (row) in TGF-beta signaling pathway from L4 to D6, which were mainly clustered into 3 groups (the group from A to C). (B) It showed the heat map and hierarchical clustering in TGF-beta signaling pathway from the stage of D6 to D15. The samples were clustered into two groups, three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together and three replicates at day 15 (D15-1, D15-2 and D15-3) were clustered together. There were also 30 involved genes in TGF-beta signaling pathway from D6 to D15, which were mainly clustered into 2 groups (the group of A and B).Red is for up-regulated and green is for down-regulated.
ErbB signaling pathway was the last one of our truly aging-dependent signaling pathways with dynamic expression pattern during aging (Figure 6). The ErbB family of receptor tyrosine kinases (RTKs) couples binding of extracellular growth factor (EGF) ligands to intracellular signaling pathways regulating diverse biologic responses, including proliferation, differentiation, cell motility, and survival [29]. In this pathway, lin-45 was the only one significantly regulated gene, which also appearing in both MAPK signaling pathway and mTOR signaling pathway (Table 2). lin-45 is a component of an EGFR-mediated inductive signaling pathway that causes vulva precursor cells (VPCs) to generate the vulva [30]. Expression of LIN-45(V627E) and LIN-45(ED) was previously identified to occasionally cause the vulva to burst at the L4-to-adult molt, which may reflect defects in either the VPC specification or the resulting vulval cells [31]. Although Hedgehog signaling pathway has been identified as one of the significantly aging-dependent pathways, it was not the true one due to the unsuccessful clustering on samples from D6 to D15 by using involved genes (Additional file 3).
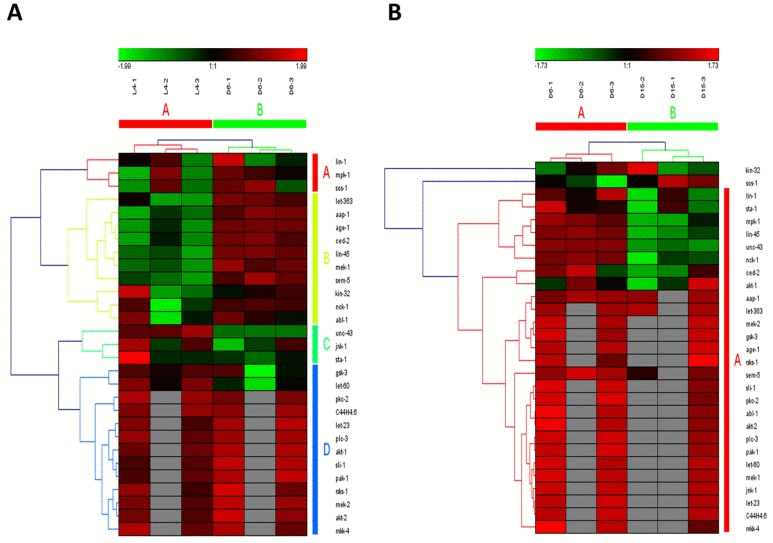
Figure 6. The heat map and hierarchical clustering in ErbB signaling pathway during aging. (A) It showed the heat map and hierarchical clustering in ErbB signaling pathway from the stage of L4 to D6. The samples (column) were clustered into two groups, three replicates in the stage of L4 (L4-1, L4-2 and L4-3) were clustered together and three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together. There were 29 involved genes (row) in ErbB signaling pathway from L4 to D6, which were clustered into 4 groups (the group from A to D). (B) It showed the heat map and hierarchical clustering in ErbB signaling pathway from the stage of D6 to D15. The samples were clustered into two groups, three replicates at day 6 (D6-1, D6-2 and D6-3) were clustered together and three replicates at day 15 (D15-1, D15-2 and D15-3) were clustered together. There were also 29 involved genes in ErbB signaling pathway from D6 to D15, in which there was mainly one group (the group of A).Red is for up-regulated and green is for down-regulated.
Generally, there were 12 significant genes with dynamic regulation patterns during aging in above five truly significant signaling pathways (Figure 7). They were ced-2, lin-45, ZK856.8, F59D6.7, pdk-1, cyd-1, sma-4, andskr-7, 8, 9, 13, 14, all of which were significantly up-regulated from L4 to D6 and down-regulated from D6 to D15. The information of significances and involved pathways were shown in Table 2. These targets would be regarded as key factors in future studies of genetic mechanisms of aging. Interestingly, 5 members of skr gene family (skr-7, 8, 9, 13, 14) were included. skr-7 encodes a homolog of Skp1 that functions within a particular SCF ubiquitin-ligase (E3) complex by binding both a cullin (a homolog of S. cerevisiae Cdc53) and an F box protein (through direct interaction with an F box motif), which is required for posterior body morphogenesis, embryonic and larval development, and cell proliferation [32, 33]. The most closely related paralogs of skr-7, -8 and -9 were highly similar, but it is not clear whether these genes comprise a functionally redundant set. The function of SKR-13 is not necessarily confined to, or even partially involved with C. elegans ubiquitin-ligase complexes. In two-hybrid assays, SKR-13 does not bind to any known C. elegans cullins (CUL-1 through CUL-6) [33]. The most closely related paralogs of skr-13 in the C. elegans genome are skr-12 and skr-14.
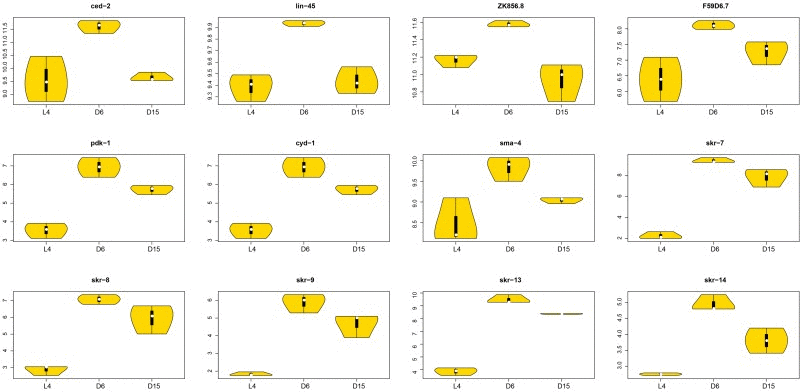
Figure 7. The violin plots of significantly associated genes in signaling pathways during aging. The violin plots showed the expression distributions of the significantly associated genes with dynamic regulation pattern in signaling pathways during aging. The X-axis represents three different stages (L4, D6 and D15); The Y-axis represents the normalized gene expression value. There were 12 genes (ced-2, lin-45, ZK856.8, F59D6.7, pdk-1, cyd-1, sma-4, and skr-7, 8, 9, 13, 14), all of which were significantly up-regulated from L4 to D6 and down-regulated from D6 to D15. The significances of each gene were shown in Table 2.
Multi-level gene regulatory networks in signaling pathways during aging
There were totally 154 and 158 individual genes in above five truly significant signaling pathways, respectively from L4 to D6 and from D6 to D15. The expression profiles of these genes were shown in Additional file 4. Based on ChIP–seq data analysis to explore the binding site targets of individual factors, 2530, 5974 and 3178 target genes were identified for transcription factors (TFs) of FOS-1, NHR-28 and UNC-62, respectively (Additional file 4). To dissect the association among these regulators or between regulators and target genes in multiple signaling pathways both from L4 to D6 and from D6 to D15, multi-level gene regulatory networks were constructed for these two developmental stages. As a result, NHR-28 was commonly identified at the top of the network, which regulated UNC-62 and FOS-1, the regulator of FOS-1 was regulated by UNC-62 at the same time (Figure 8). HOX co-factor UNC-62 (Homothorax) has been identified as a developmental regulator that binds proximal to age-regulated genes and modulates lifespan by integrating RNAi and genomics approach [39]. C. elegans FOS-1 has been shown to act in uterine and vulval development, which regulates plc-1 expression in the spermatheca to control ovulation [4]. Moreover, 64 interactions between regulatory factors and target genes were identified at the first stage, including 26 genes regulated by NHR-28, 21 genes by UNC-62 and 17 genes by FOS-1. By contrast, 79 interactions between regulatory factors and target genes were identified at the second stage, including 31 genes regulated by NHR-28, 27 genes by UNC-62 and 21 genes by FOS-1 (Additional file 5). 32 common target genes were identified at both stages, but 3 genes only appears at the first stage (ruvb-1, let-60 and siah-1 in Figure 8A) and 10 genes only appears at the second stage (cdc-42, kin-19, mom-5,ife-2, mek-2, C07E3.9, gpa-12, sos-1, let-23and skr-21 in Figure 8B).
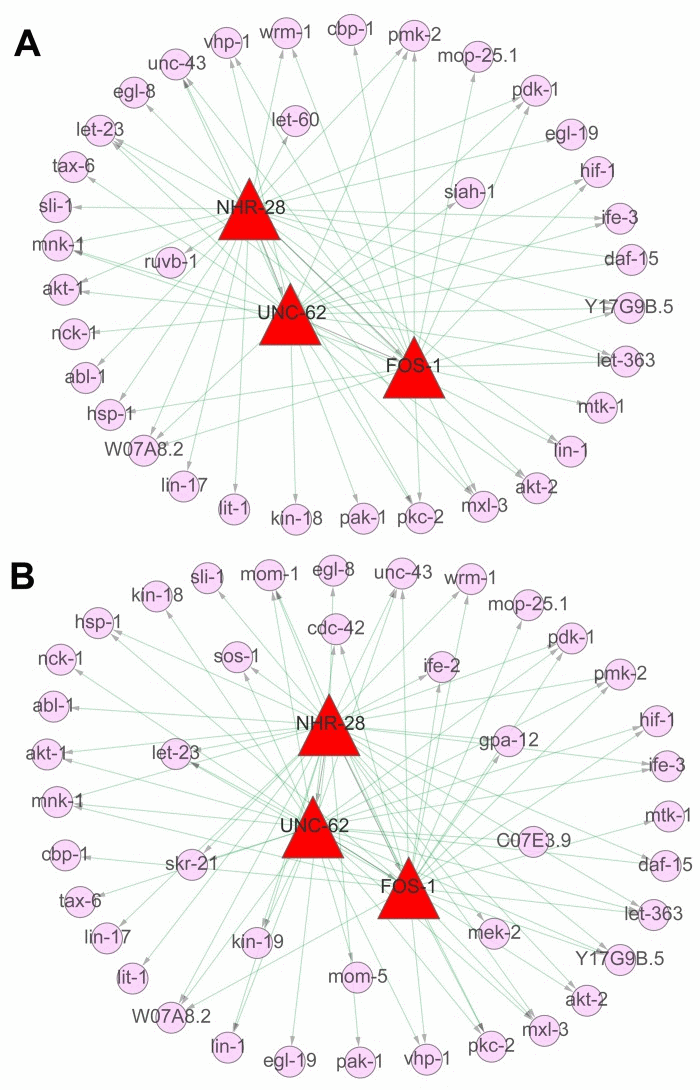
Figure 8. The regulatory networks in signaling pathways during aging. The regulatory networks were constructed based on three of aging related ChIP–seq datasets (3 TFs of NHR-28, UNC-62 and FOS-1 related) and the expression matrices of 154 involved genes in identified signaling pathways above. (A) It indicated the gene regulatory networks from the stage of L4 to D6, including the TFs (NHR-28, UNC-62 and FOS-1 in red triangular frame) as well as the target genes (in pink circle). (B) It indicated the gene regulatory networks from the stage of D6 to D15, including the TFs (NHR-28, UNC-62 and FOS-1 in red triangular frame) as well as the target genes (in pink circle). The external part represents the common genes between these two periods and the inner part represents the novel targets not appearing in the other period.
In this study, we employed the gene expression datasets of three time points during aging in C. elegans and performed the approach of GSEA on each dataset between two adjacent stages. As a result, multiple genetic pathways and targets were identified as significantly down- or up-regulated. Among them, 5 truly aging-dependent signaling pathways including MAPK signaling pathway, mTOR signaling pathway, Wnt signaling pathway, TGF-beta signaling pathway and ErbB signaling pathway as well as 12 significantly associated genes were identified with dynamic expression pattern during aging. On the other hand, the continued declines in the regulation of several metabolic pathways have been demonstrated to display age-related changes. Furthermore, the reconstructed regulatory networks based on three of aging related ChIP–seq datasets and the expression matrices of 154 involved genes in above signaling pathways provided new insights into aging at the multiple pathways level. In conclusion, the identification of these genetic pathways and target genes by our integrated analysis would support the hyperfunction theory, which is a plausible alternative to the molecular damage theory to explain aging in C. elegans [1]. The combination of multiple genetic pathways and targets needs to taken into consideration in future studies of aging, in which the dynamic regulation would be uncovered.