High throughput microarray detection of plasma lncRNAs and microRNAs
In total, 597 patients diagnosed with CRC, 585 paired healthy controls, and 19 patients diagnosed with CRA were enrolled. All participants in this study was age and gender matched. For the CRC patients, the subgroup was divided according to the Differentiation grade, tumor size (with 5cm as cutoff), with or without metastasis, and tumor TNM staging. The detailed clinical information was presented in Table 1.
Table 1. Clinicopathological features of surgical colorectal cancer (CRC) and cancer-free control samples.
| CRC | CRA | Control | P valve |
| N | 597 | 19 | 585 | |
| Age Mean (SE) year | 62.89(0.02) | 57.32(0.63) | 57.17(0.02) | 0.32a |
| Sex (male/female) | 376/221 | 10/9 | 357/228 | 0.55b |
| Differentiation grade | | | | |
| Well | 0 | | | |
| Moderate | 373 | | | |
| Poorly | 224 | | | |
| Tumor Size(cm) | | | | |
| ≤5 cm | 427 | | | |
| >5 cm | 170 | | | |
| Metastasis | | | | |
| Yes | 288 | | | |
| No | 309 | | | |
| Tumor stage | | | | |
| Stage I, II | 309 | | | |
| Stage III, IV | 288 | | | |
| TNM staging system | | | | |
| T1+T2 | 156 | | | |
| T3+T4 | 441 | | | |
| a Student t-test. |
| b Chi-square test. |
First, plasma RNA was extracted from CRC group, CRA group and Control group. Samples were applied to the miRNA and lncRNA microarray. Each group we enrolled three samples. Hierarchical clustering analysis and volcano plot distribution were used to sort the aberrantly expressed miRNAs/lncRNAs in different groups. As presented in Figure 1A and 1B, different expression level of miRNA and lncRNA in each group were obtained. Then further screening was performed as follows: a, P value <0.05; b, CT value <35; c, detection rate >75%. Total of 79 miRNA transcripts were specifically increased in CRA group comparing with NC group, 105 miRNAs were collected in CRC group by comparing with CRA group. In order to screen the biomarker for predicating, the Venny analysis was applied and finally yielded 6 miRNAs candidates as listed in Figure 1C and 1E. For lncRNA, total of 185 lncRNA transcripts were specifically increased in CRA group comparing with NC group, 274 lncRNAs were collected in CRC group by comparing with CRA group. The Venny analysis finally yielded 6 lncRNAs candidates as listed in Figure 1D and 1F.
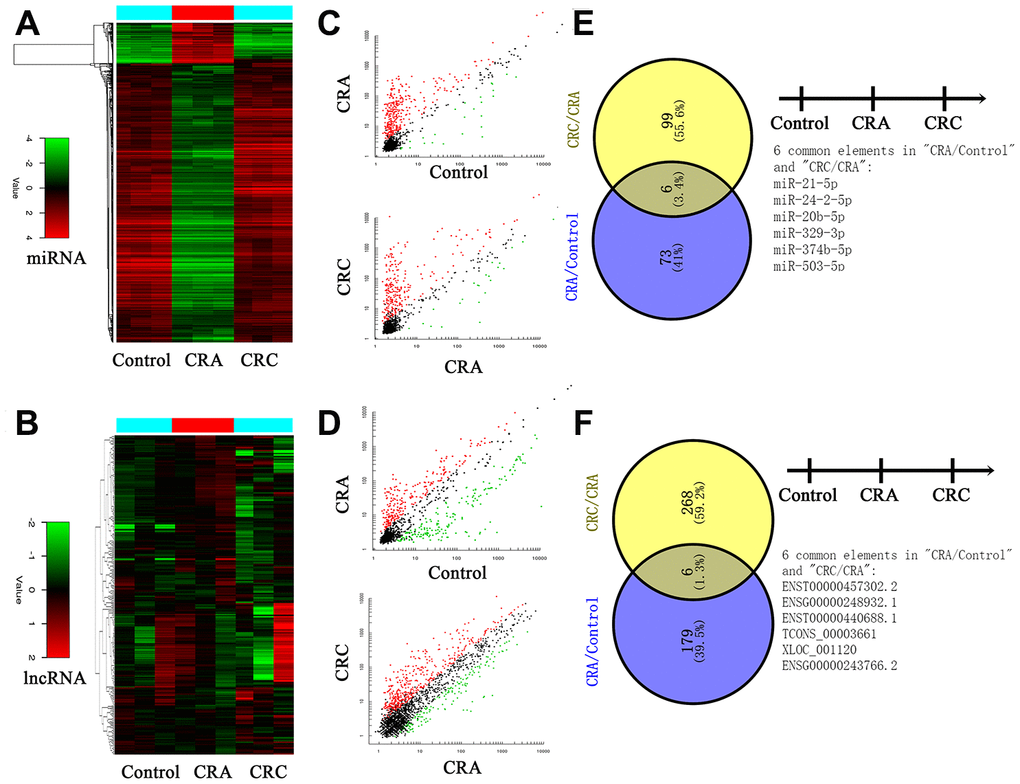
Figure 1. Circulating non-coding RNA expression landscape of in HC, CRA and CRC patients. (A, B) Cluster analysis for the miRNA and lncRNA expression in HC, CRA and CRC groups. Each group including three samples. (C, D) The scatter distribution of aberrant expressed miRNA/lncRNA in different groups. (E, F) The candidate miRNA/lncRNA was screened through Venny analysis.
Next, a larger sample scale was employed for further validation the 12 candidates. As presented in Figure 2, among the 12 miRNA/lncRNA, one of which entitled with ENST00000457302.2 presented no amplification with the RT-PCR assay. Two miRNAs including miR-21-5p and miR-24-2-5p, three lncRNA including ENSG00000248932.1, ENST00000440688.1 and TCONS_00003661 presented no difference. Therefore, a panel of 6 non-coding RNAs including miR-20b-5p, miR-329-3p, miR-374b-5p, miR-503-5p, XLOC_001120 and ENSG00000243766.2 were selected the further validation analysis.
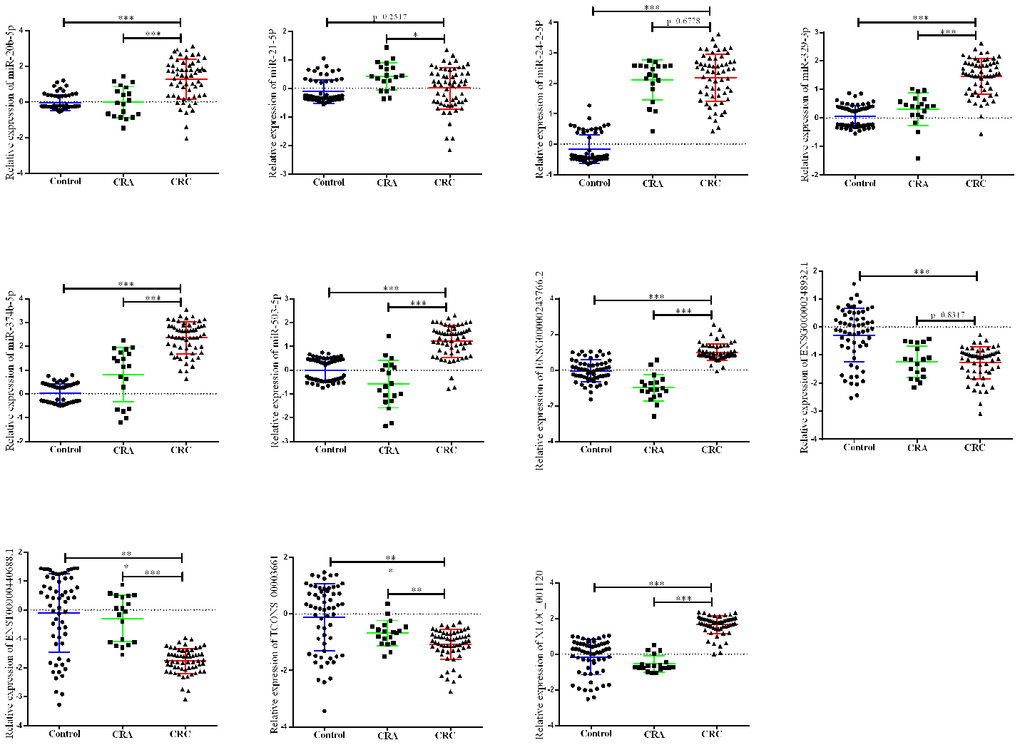
Figure 2. Relative expression of candidate non-coding RNA through first-phage validation. qRT-PCR analysis was used to detect the expression of 6 miRAN and 6 lncRNA in 40 paired plasma samples from healthy controls, 19 samples of CRA patients and 40 plasma samples from CRC patients. Data was log-transformed and was presented as mean ± SD. Data was analyzed with student t test. “***” indicated p < 0.001.
Training set and validation set for selecting the biomarker for CRC diagnosis
The panel of 6 non-coding RNAs was found to be effective markers for the diagnosis of CRC through the abovementioned experimental design by using multiphase detection and analysis. The expression of miR-20b-5p, miR-329-3p, miR-374b-5p, miR-503-5p (Figure 3A–3D, Supplementary Tables 1 and 2) and lncRNA including XLOC_001120 and ENSG00000243766.2 were significantly increased in the CRC plasma samples compared with CRA and healthy control plasma samples (Figure 4A and 4B, Supplementary Tables 1 and 2). In addition, we also detected relative expression of miR-20b-5p, miR-329-3p, miR-374b-5p, miR-503-5p, XLOC_001120 and ENSG00000243766.2 through qRT-PCR in 60 pairs CRC tissues and matched adjacent tissues. All 6 non-coding RNAs were increased in the CRC tissues (Supplementary Figure 1A–1F).
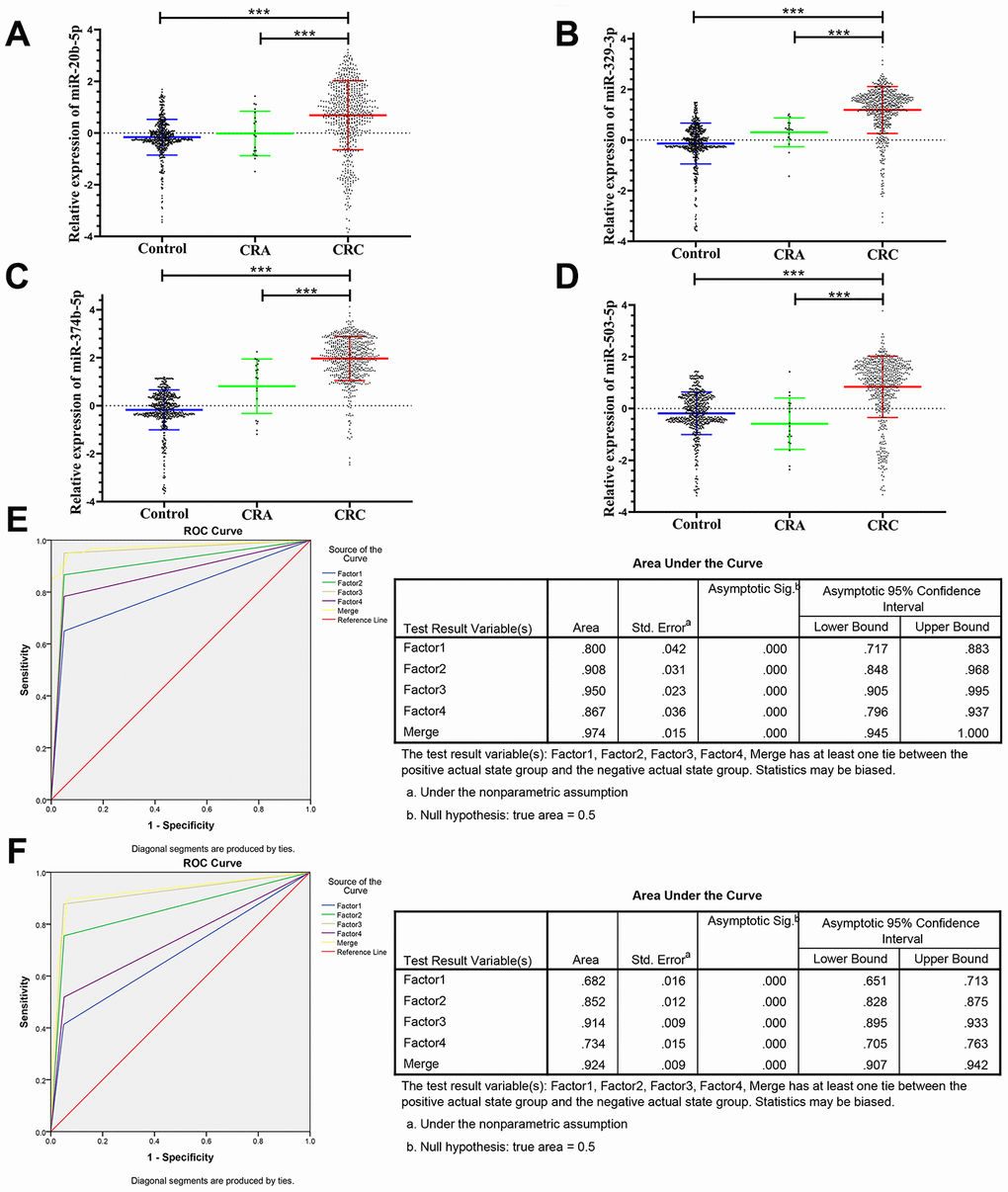
Figure 3. Relative expression of 4 microRNAs in HC, CRA and CRC, and ROC curve analysis for predicting the 4 microRNAs as CRC diagnosis biomarkers. (A–D) qRT-PCR analysis was used to detect the expression of miR-20b-5p, miR-329-3p, miR-374b-5p and miR-503-5p in 585 plasma samples from healthy controls, 19 samples of CRA patients and 597 plasma samples from CRC patients. Data was log-transformed and was presented as mean ± SD. Data was analyzed with student t test. “***” indicated p < 0.001. (E) ROC curve for the 4-microRNA signature to separate 60 CRC cases from 60 controls in the training set with the AUC presented in the right. (F) ROC curve analysis was used for the 4-microRNA signature to differentiate 597 CRC cases from 585 controls in the validation set with the AUC presented in the right. Factor1, 2, 3, 4 and merged represented the miR-20b-5p, miR-329-3p, miR-374b-5p, miR-503-5p and the combination of the 4 microRNAs.
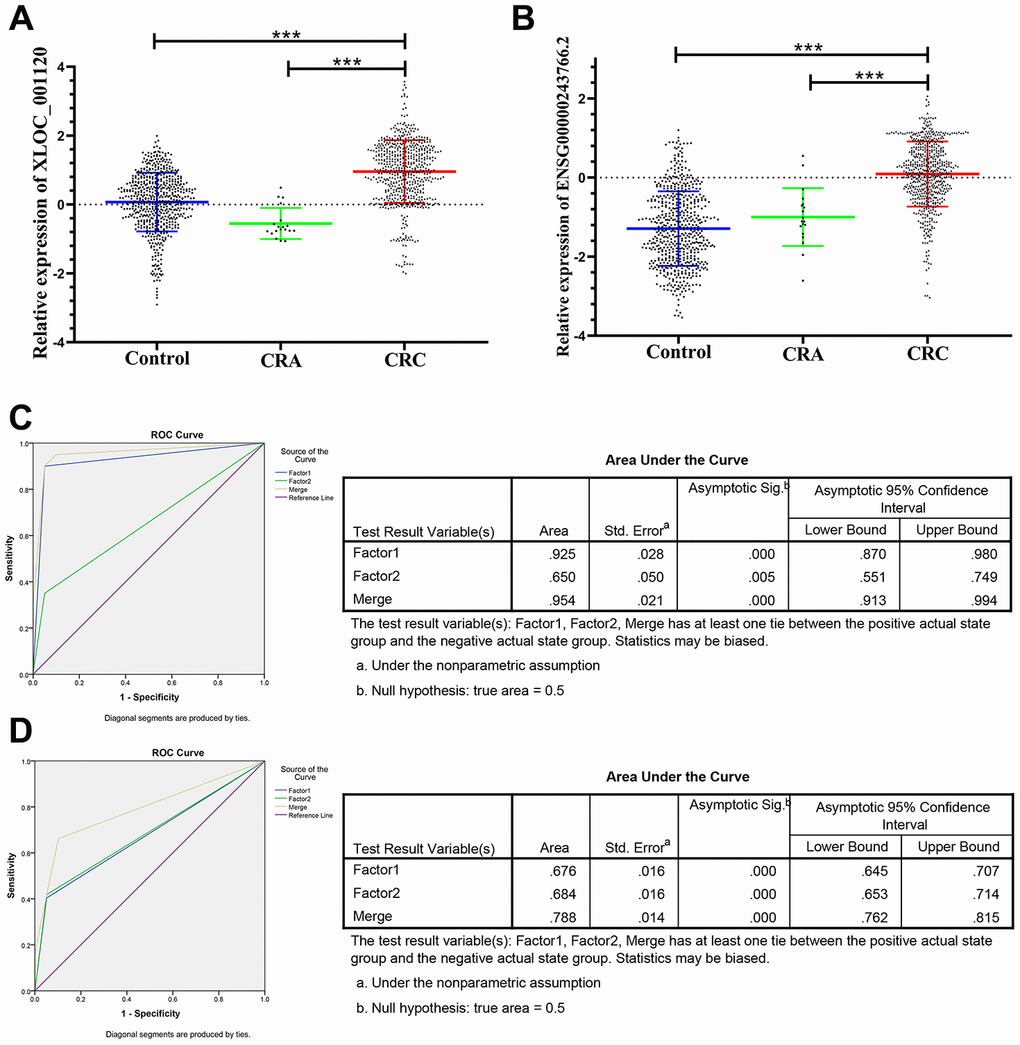
Figure 4. Relative expression of 2 lncRNAs in HC, CRA and CRC, and ROC curve analysis for predicting the 2 lncRNAs as CRC diagnosis biomarkers. (A–B) qRT-PCR analysis was used to detect the expression of XLOC_001120 and ENSG00000243766.2 in 585 plasma samples from healthy controls, 19 samples of CRA patients and 597 plasma samples from CRC patients. Data was log-transformed and was presented as mean ± SD. Data was analyzed with student t test. “***” indicated p < 0.001. (C) ROC curve for the 2-lncRNA signature to separate 60 CRC cases from 60 controls in the training set with the AUC presented in the right. (D) ROC curve analysis was used for the 2-lncRNA signature to differentiate 597 CRC cases from 585 controls in the validation set with the AUC presented in the right. Factor1, 2 and merged represented the XLOC_001120, ENSG00000243766.2 and the combination of the 2 lncRNAs.
Risk score analysis (RSA) was used to evaluate the predicting ability of the panel of 6 non-coding RNAs as CRC diagnostic markers. First, the risk score of each plasma sample were calculated and taken as a parameter for further logistic regression model. The calculated cutoff of risk score was used to divide the plasma sample into the high score group (representing predicted CRC) and the low score group (representing possible cancer-free group). Combined sensitivity and specificity were maximized at a cut-off score of 9.825, and the prediction accuracy of CRC and prediction value of cancer-free control was 0.97 and 0.97 in the training set, respectively. Then, verification of the effectiveness under the cutoff value in the larger validation samples showed the positive predictive value and negative predictive value was 0.96 and 0.77, respectively (Table 2).
Table 2. Risk score analysis of CRC and cancer-free control plasma samples.
| Score | 0–9.825 | 9.825–19.65 | PPV | NPV |
| Training set | | | 0.97 | 0.97 |
| CRC | 2 | 58 | | |
| Control | 58 | 2 | | |
| Validation set | | | 0.96 | 0.77 |
| CRC | 170 | 427 | | |
| Control | 569 | 16 | | |
| PPV, positive predictive value. |
| NPV, negative predictive value. |
The ROC analysis was used to evaluate the diagnostic performance of the chose non-coding RNAs panel by using risk score analysis. As shown in Figures 3E, 4C and 5A, the area under the curve (AUC) of miR-20b-5p, miR-329-3p, miR-374b-5p, miR-503-5p, XLOC_001120, ENSG00000243766.2 and their combination was 0.800, 0.908, 0.950, 0.867, 0.925, 0.650 and 0.996 in training set. When the sample size expanded to 597 CRC vs 585 HC, the AUC for the non-coding RNAs and their combination was 0.682, 0.852, 0.914, 0.734, 0.676, 0.684 and 0.954 respectively (Figures 3F, 4D and 5B).
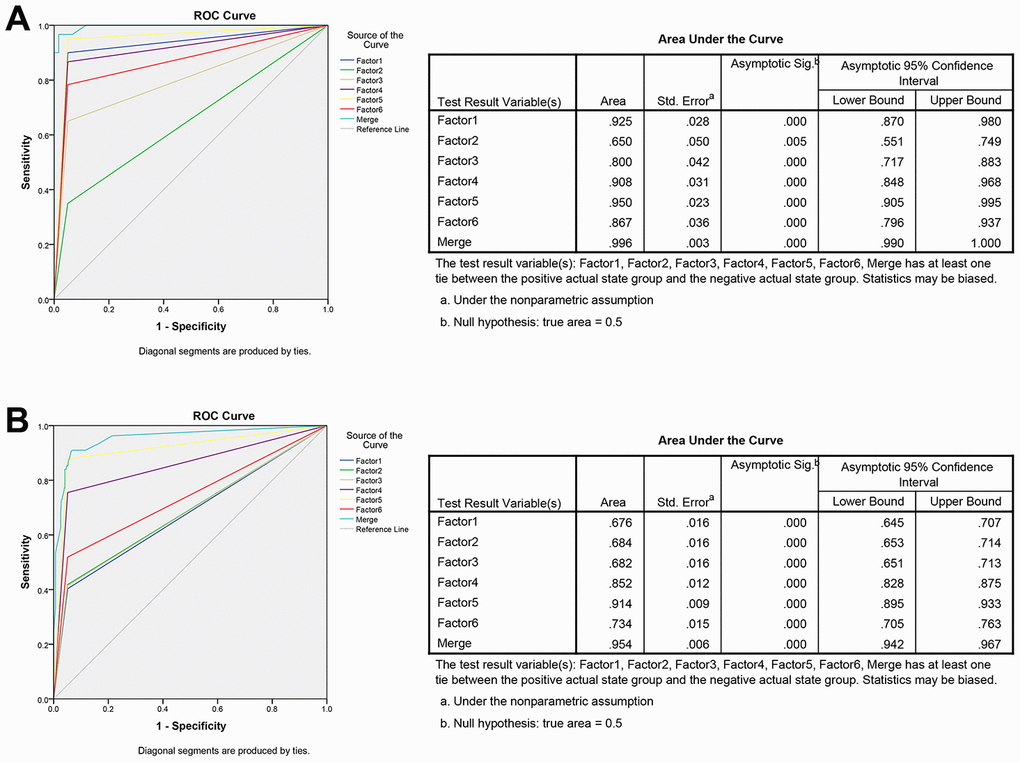
Figure 5. ROC curve analysis for predicting the 6 non-coding RNAs as CRC diagnosis biomarkers. (A) ROC curve for the 6 non-coding RNAs signature to separate 60 CRC cases from 60 controls in the training set with the AUC presented in the right. (B) ROC curve analysis was used for the 6 non-coding RNAs signature to differentiate 597 CRC cases from 585 controls in the validation set with the AUC presented in the right. Factor1, 2, 3, 4, 5, 6 and merged represented the XLOC_001120, ENSG00000243766.2, miR-20b-5p, miR-329-3p, miR-374b-5p, miR-503-5p and the combination of the 6 non-coding RNAs.
The panel of miR-20b-5p, miR-329-3p, miR-374b-5p, miR-503-5p, XLOC_001120 and ENSG00000243766.2 was used to differentiate the CRC and CRA by using similar risk score analysis and ROC analysis. The expression of these 6 non-coding RNAs was significantly increased in CRC plasma samples compared with the CRA plasma samples (Supplementary Table 2). The AUC of miR-20b-5p, miR-329-3p, miR-374b-5p, miR-503-5p and their combination was 0.874, 0.924, 0.861, 0.799 and 0.939 in training set, and was 0.645, 0.838, 0.713, 0.715 and 0.850 in the 597 CRC samples vs 19 CRA samples, respectively (Supplementary Figure 2C, 2D). As shown in Supplementary Figure 2A, 2B, The AUC of XLOC_001120, ENSG00000243766.2 and their combination was 0.749, 0.736 and 0.818 in the 40 CRC samples vs 19 CRA samples, and was 0.827, 0.614 and 0.869 in the validation set, respectively. A repeated validation test in the independent datasets indicated that the expression of lncRNAs and microRNAs only elevated in the plasma of CRC patients not in the CRA patients and healthy people (Supplementary Figure 5, Supplementary Table 3).
miR-20b-5p, miR-329-3p and miR-503-5p acting as the tumor size indicator via clinicopathological relevance analysis for CRC
Previous studies reported that the clinicopathological characteristics (including tumor size, differentiation grade, and metastasis) were significantly associated with the progression and prognosis of CRC [3, 26]. Therefore, we further analyzed the expression levels of the 6 non-coding RNAs in three following subgroups (tumor size, differentiation grade and metastasis) that based on the 597 CRC plasma samples. The results showed that there was no significant difference regarding to the tumor differentiation (well, medium or poor) and metastasis (with or without) (Supplementary Figure 3). However, 3 of the 6 non-coding RNAs, miR-20b-5p, miR-329-3p and miR-503-5p significantly elevated in plasma samples from larger tumors (5 cm as the cutoff) in CRC patients (Figure 6A).
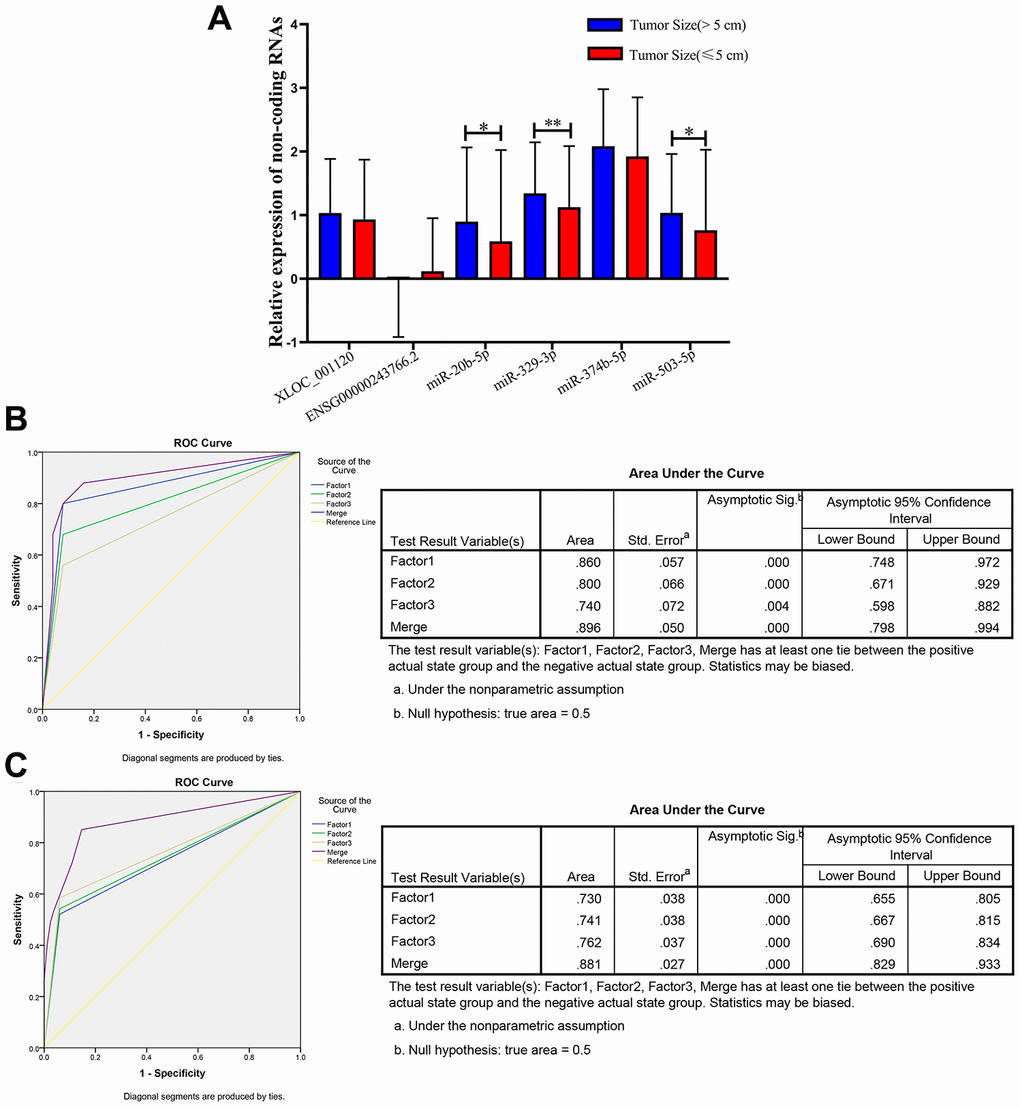
Figure 6. Relative expression of 6 non-coding RNAs in different tumor size of CRC, ROC curve analysis for predicting 3 microRNAs as a CRC tumor size biomarker. (A) qRT-PCR analysis was used to detect the expression of XLOC_001120, ENSG00000243766.2, miR-20b-5p, miR-329-3p, miR-374b-5p and miR-503-5p in 170 plasma samples from larger size (size>5cm) CRC patients and 427 smaller size (size≤5cm) CRC patients. Data was log-transformed and was presented as mean ± SD. Data was analyzed with student t test. “*” indicated p < 0.05, “**” indicated p < 0.01. (B) ROC curve analysis was conducted to discriminate between larger size group and smaller size group by the 3-microRNA profile. ROC curve analysis was performed for the 3-microRNA signature to separate 25 pairs in the training set with the AUC presented in the right. (C) ROC curve analysis was used for the 3-microRNA signature to differentiate 94 larger size CRC cases from 82 smaller size CRC group in the validation set with the AUC presented in the right. Factor1, 2, 3 and merged represented the miR-20b-5p, miR-329-3p, miR-503-5p and the combination of the 3 microRNAs.
Therefore, we randomly selected 25 (tumor size>5cm)/ 25 (tumor size≤5cm), 94 (tumor size>5cm)/ 82 (tumor size≤5cm) plasma samples as the training set and validation set of CRC to further investigate the diagnostic efficiency of miR-20b-5p, miR-329-3p and miR-503-5p. The elevated expression levels of 3 microRNAs were confirmed in the training set and validation set (Supplementary Table 4). The sensitivity and specificity of microRNAs for diagnosing larger tumor size were 94% and 75% in the training set with cutoff value 4.40, respectively. In addition, the same cutoff value was used to calculate the risk score of the validation set samples. The diagnostic sensitivity was 94%, the specificity was 64% (Table 3). The AUC of miR-20b-5p, miR-329-3p and miR-503-5p was 0.86, 0.8, 0.74 and the combination was 0.896 in training set. The AUCs in validation set were 0.73, 0.741, 0.762 and 0.881, respectively. The results indicated that the panel of three microRNAs may be a novel biomarker of diagnosis larger CRC tumor (Figure 6B and 6C).
Table 3. Risk score analysis of tumor size in CRC patients’ plasma samples.
| Score | 0–4.40 | 4.40–8.80 | PPV | NPV |
| Training set | | | 0.94 | 0.75 |
| Size(>5 cm) | 8 | 17 | | |
| Size(≤5 cm) | 24 | 1 | | |
| Validation set | | | 0.94 | 0.64 |
| Size(>5 cm) | 44 | 50 | | |
| Size(≤5 cm) | 79 | 3 | | |
| PPV, positive predictive value. |
| NPV, negative predictive value. |