PSMC family members play crucial roles in breast cancer progression
Previous studies identified six PSMC family members in Homo species, and some of these genes play crucial roles in cancer progression. Oncomine platform contained a total of 392 unique analyses for PSMC1 expression, and PSMC1 had significant in 13 of 392 unique analyses. PSMC2 had significant in 55 of 433 unique analyses, PSMC3 had significant in 28 of 421 unique analyses, PSMC4 had significant in 71 of 432 unique analyses, PSMC5 had significant in 24 of 420 unique analyses, PSMC6 had significant in 28 of 445 unique analyses (Figure 1). However, a meta-analysis is needed to clarify gene signatures of PSMC family members in breast cancer. According to our results from an Oncomine analysis of mRNA expressions of PSMC2, PSMC3, PSMC4, PSMC5, and PSMC6, these members are highly upregulated in breast cancer tissues; therefore, we chose breast cancer to perform further bioinformatics analyses (Figure 1). Furthermore, in the METABRIC database, expressions of PSMC members in breast cancer tissues were significantly higher than those in normal tissues; p values ranged from 2.16E-45 to 0.023 for PSMC1, 1.37E-29 to 0.016 for PSMC2, 3.18E-21 to 0.001 for PSMC3, 1.28E-53 to 0.018 for PSMC4, 7.02E-36 to 0.041 for PSMC5, and 9.03E-12 to 0.039 for PSMC6 (Supplementary Table 1). Meanwhile, to further explore gene expressions of the entire PSMC family in breast cancer, we compared transcript levels of different breast cancer subtypes, such as the triple-negative, HER-2, and luminal subtypes, relative to normal breast tissues, in TCGA database (Supplementary Figure 1). Interestingly, we discovered that PSMC genes were overexpressed in a subtype-specific manner: specifically, PSMC1, PSMC2, PSMC3, PSMC4 were highly expressed in the triple-negative subtype, PSMC5 in HER-2, and PSMC6 in luminal cancer. These results suggest oncogenic effects of PSMC family genes on tumor progression.
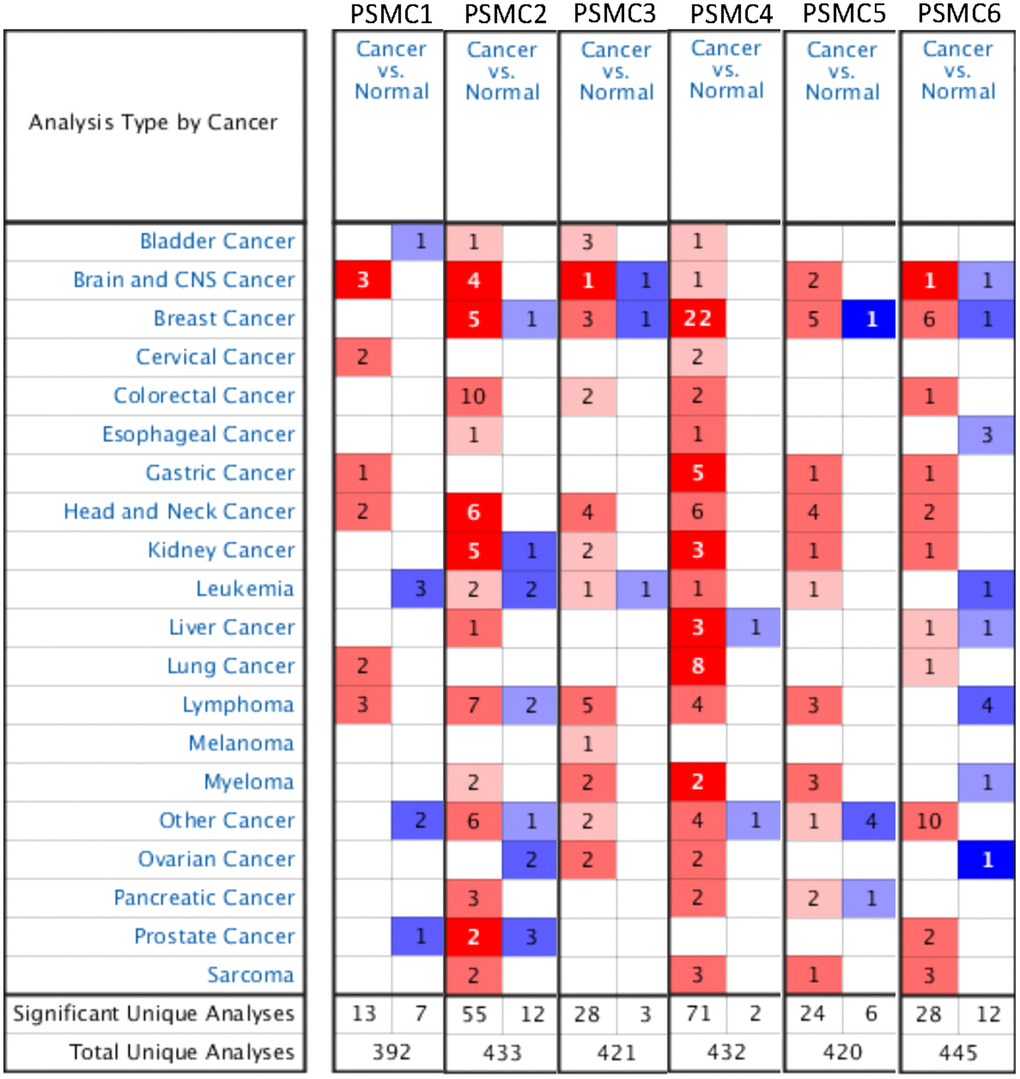
Figure 1. Overview of mRNA expression levels of proteasome 26S subunit, ATPase (PSMC) genes in multiple types and subtypes of cancer from the Oncomine database. The analysis compared expressions of target genes in breast cancer tissues relative to normal matched tissues. Red and blue gradients display the top-ranked genes in specific datasets. Significant unique analysis means the number of datasets that satisfied the threshold of >2 multiples of change, p<0.05, and in the top 10% gene ranking.
Genes coexpressed with PSMC family genes in breast cancer
We analyzed genes coexpressed with PSMC1 in the Perou Breast 2 dataset from the Oncomine platform. We found that PSMC1 was positively correlated with MYL9, PCOLCE, ANXA6, DVL1, MAPKAPK2, PROC, FTL, ZNF358, CHRNA1, COL5A1, MPP1, PDE6A, COL1A1, MIA, SH2B3, COL6A1, and BGN. We used the Landemaine dataset to analyze genes coexpressed with PSMC2 and found that its expression was positively correlated with SMAD5, KLHL20, ZNF148, KLHL28, PPP4R2, SFRS12IP1, NPHP3, TMCC1, KIAA2018, DHX29, Clorf27, C14orf138, SOCS4, Cllorf46, FKTN, RNF170, CHIC1, ZNHIT6, and JMIDIC. We analyzed genes coexpressed with PSMC3 in the Minn dataset and found that its expression was positively correlated with EIF6, CLIC1, MAPRE1, EIF2S2, GRPEL1, TMEM93, PSMB6, EXOSC9, RPAI, COPS3, G3BP1, EIF2S1, NOL7, SNRPC, EEFIEl, RDBP, and CSEIL. As for the genes coexpressed with PSMC4, we used the Ma dataset and found that it was positively correlated with expressions of ADRM1, CAPZB, ACOT7, HSPB1, UEVLD, PCBD2, CCDC64, C7orf68, SCD, CYB561, GPRC5A, DNAJA4, HAGH, SNRNP25, PSMD2, ANXA4, GRB2, UBE2F, and UBEZF. We analyzed genes coexpressed with PSMC5 in the Julka dataset and found that its expression was positively correlated with CCDC45, CMYA5, KCTD3, SPOPL, TP53INP1, TPPP3, C20orf54, PTGER3, EXOC1, CAT, WDR11, SDCBP, CCDC46, C20orf3, PLK1S1, MYADM, ADAMTSL3, ABCC5, and CAPS. For genes coexpressed with PSMC6, we used the Kreike dataset and found that its expression was positively correlated with CLPX, CCDC9OB, FAM18B2, C60rf62, ZBTB33, PYROXD1, CDC42SE2, COMMD6, LOC401397, CAPZAI, TPRKB, GABPA, MATR3, ZDHHC20, SCOC, and COPS2 (Figure 4A).
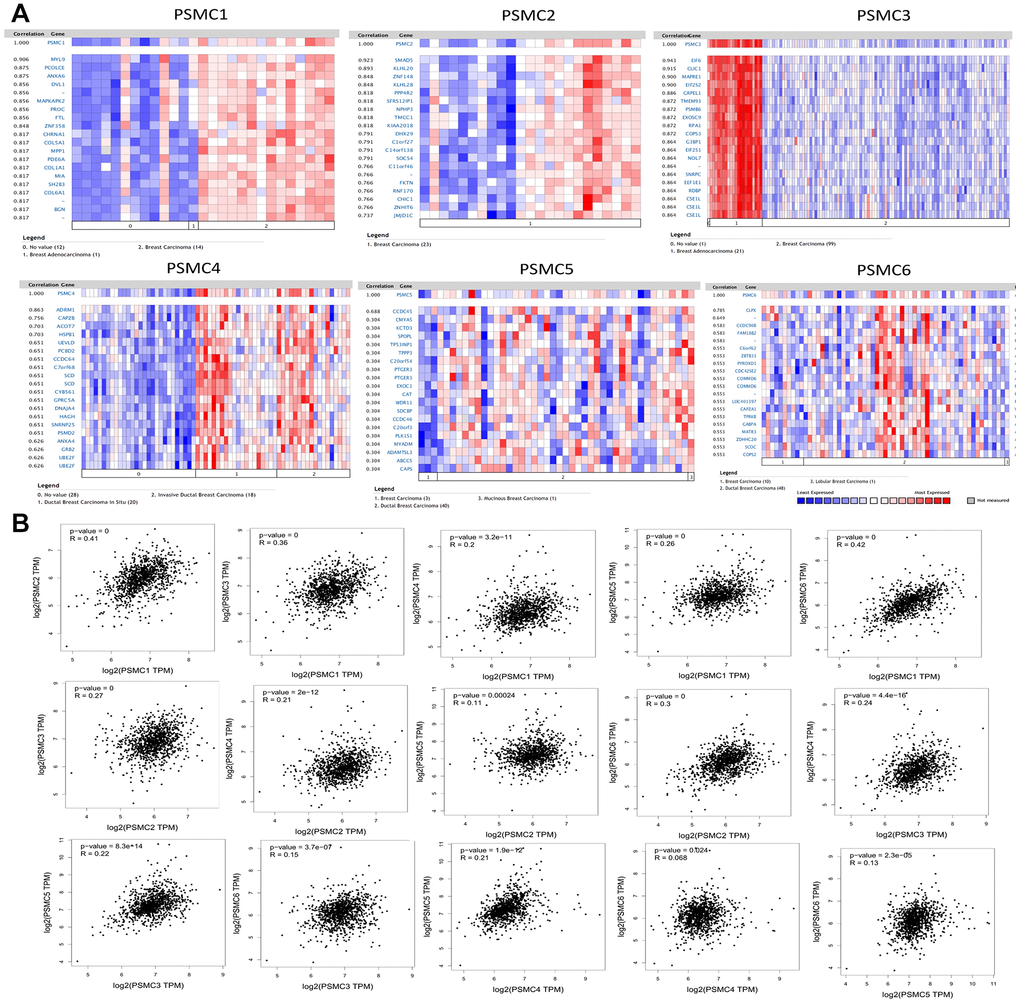
Figure 4. Genes coexpressed with the proteasome 26S subunit, ATPase (PSMC) family and correlations among the six PSMC genes in breast cancer. (A) Genes coexpressed with PSMC genes in breast cancer from the Oncomine platform. (B) Correlations among PSMC genes in breast cancer from the GEPIA2 platform.
Additionally, associations among PSMC1, PSMC2, PSMC3, PSMC4, PSMC5, and PSMC6 were also analyzed using the GEPIA dataset. Specifically, PSMC1 was positively correlated with PSMC2 (R=0.41, p<0.05), PSMC3 (R=0.36, p<0.05), PSMC4 (R=0.2, p<0.05), PSMC5 (R=0.26, p<0.05), and PSMC6 (R=0.42, p<0.05). PSMC2 was positively correlated with PSMC3 (R=0.27, p<0.05), PSMC4 (R=0.21, p<0.05), PSMC5 (R=0.11, p<0.05), and PSMC6 (R=0.3, p<0.05). PSMC3 was positively correlated with PSMC4 (R=0.24, p<0.05), PSMC5 (R=0.22, p<0.05), and PSMC6 (R=0.15, p<0.05). PSMC4 was positively correlated with PSMC5 (R=0.021, p<0.05) and PSMC6 (R=0.068, p<0.05). Finally, PSMC5 was positively correlated with PSMC6 (R=0.13, p<0.05) (Figure 4B). Meanwhile, we obtained similar results from the cBioPortal and the Cytoscaped and METABRIC databases, which revealed that the six PSMC members were correlated with cell cycle-related genes (Supplementary Figure 2). In addition, expressions of PSMC family members were also correlated with immune infiltration profiles in breast cancer, as analyzed with the Tumor Immune Estimation Resource (TIMER; cistrome.shinyapps.io/timer) tool. Expression of each PSMC gene was associated with tumor purity and markers of different types of immune cells (Supplementary Figure 3).
Protein expressions and prognostic values of the PSMC family in breast cancer specimens
Since PSMC family genes were differentially expressed in samples from breast cancer patients, we next explored the potential roles of these genes in human breast cancer tissues, correlating their expressions with other potential biomarkers related to molecular subtypes of breast cancer. To determine expressions of PSMC family members and their clinical relevance, we analyzed protein expressions of individual PSMC members in clinical specimens from the Human Protein Atlas. The data demonstrated that PSMC1-6 presented moderate protein expressions, and PSMC2, PSMC3, and PSMC5 were highly expressed in certain clinical tissues from breast cancer specimens (Figure 5). The Kaplan–Meier plotter database also showed that PSMC1, PSMC3, PSMC4, PSMC5, and PSMC6 had high expression levels in breast cancer tissues may have oncogenic roles in breast cancer progression. High transcription levels of PSMC1, PSMC3, PSMC4, PSMC5, and PSMC6 predicted poor survival, whereas PSMC2 did not show the same pattern (Figure 6).
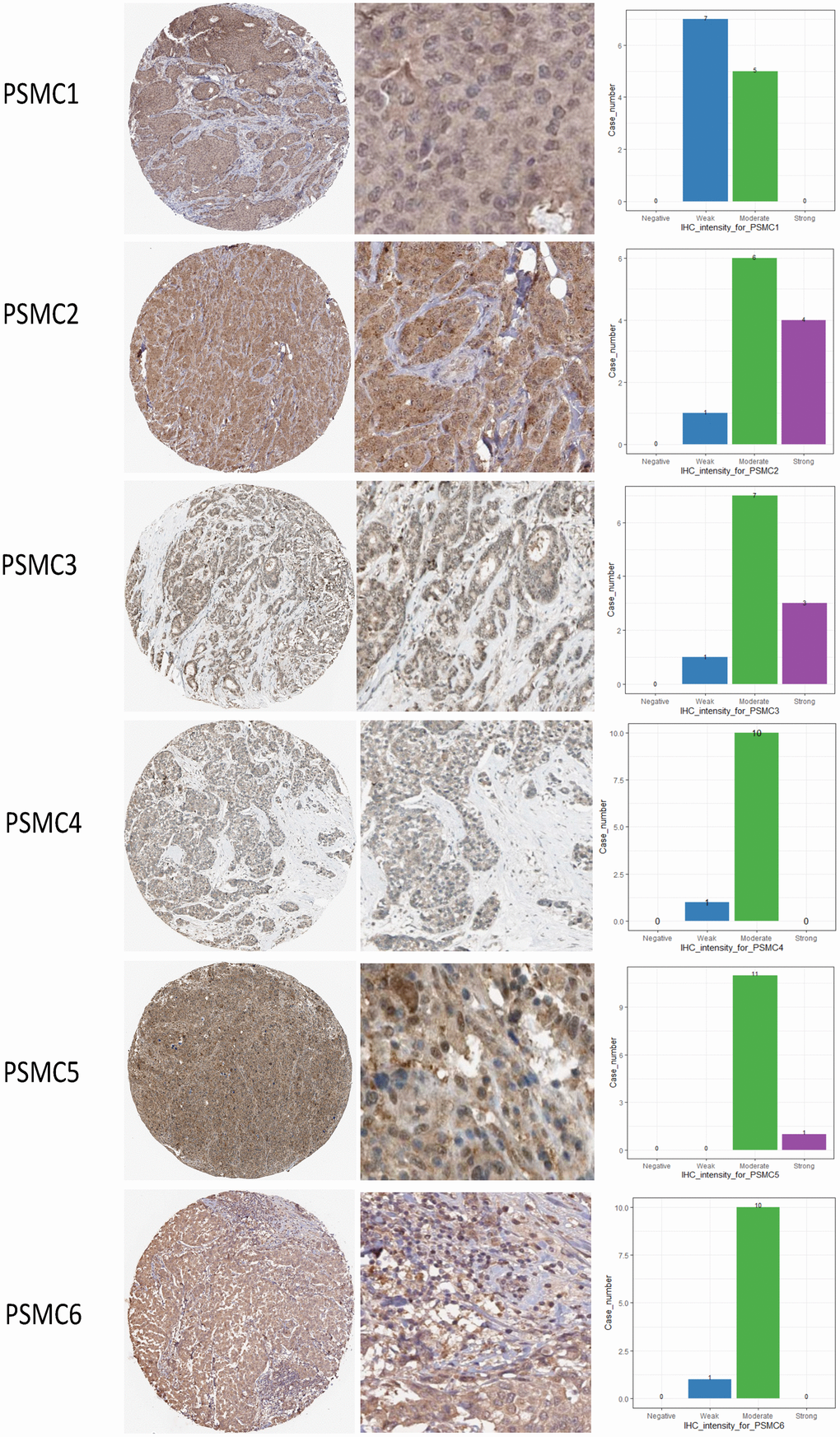
Figure 5. Protein expression levels of proteasome 26S subunit, ATPase (PSMC) family members across clinical specimens of breast cancer. PSMC1, PSMC4, and PSMC6 proteins were moderately expressed, and some clinical tissues showed strong PSMC2, PSMC3, and PSMC5 protein expressions in breast cancer.
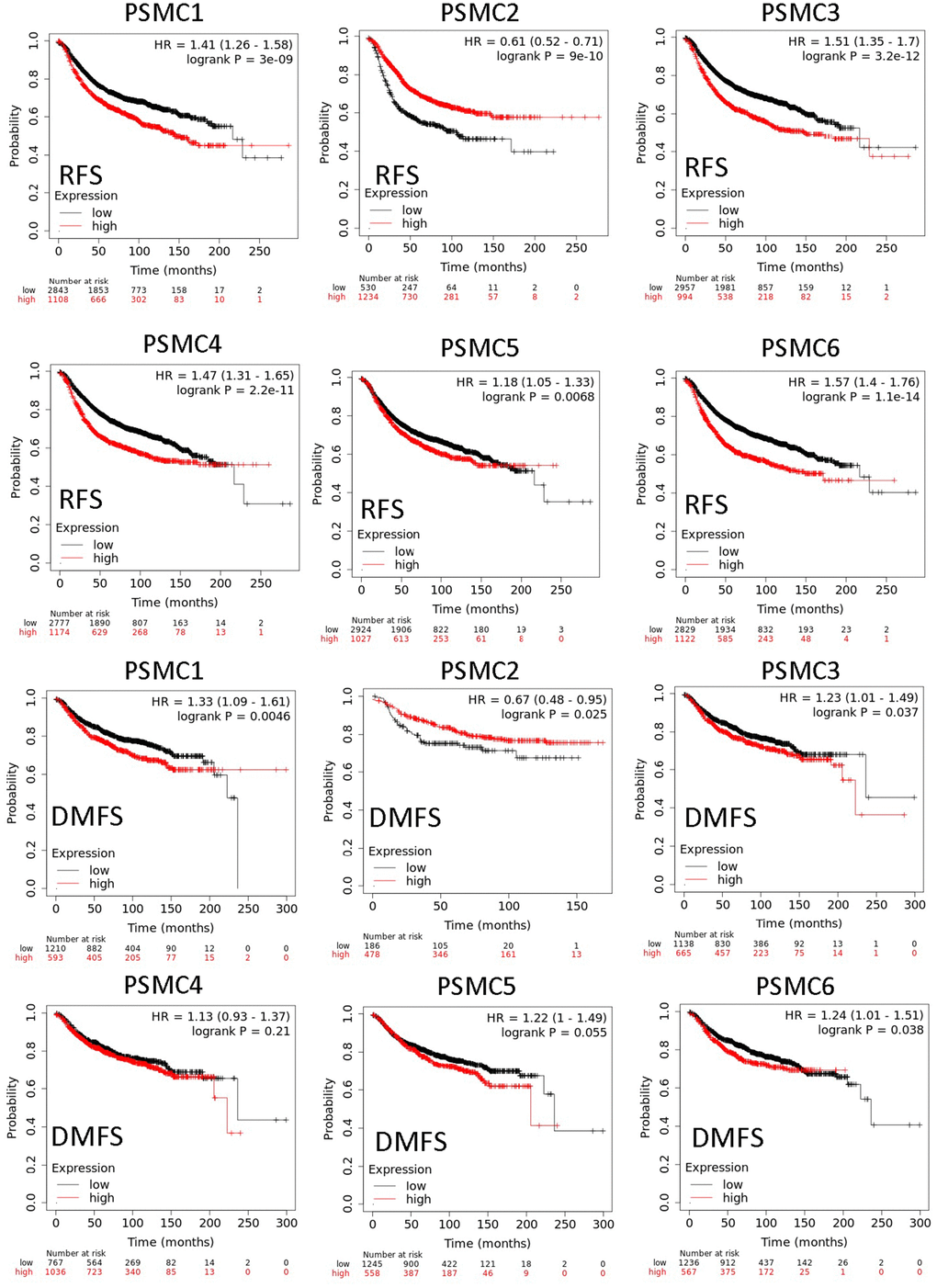
Figure 6. Relationship between expressions of proteasome 26S subunit, ATPase (PSMC) family members with recurrence-free survival (RFS) and distant metastasis-free survival (DMFS) from clinical breast cancer patients (n=2898). Kaplan–Meier plots show correlations of RFS and DMFS in breast cancer patients with high and low expression levels of PSMC family members using the median of expression as the cutoff. Red and black lines respectively represent higher and lower values than the median. High expression levels of most PSMC members were associated with poor survival, whereas high expression levels of PSMC2 were associated with significantly better survival rates (p<0.05).
Pathway and network analyses of PSMC family member genes
First, to explore the universally regulated pathways of the entire PSMC family, GeneGo Metacore was leveraged to investigate downstream networks according to coexpression patterns of PSMC genes. An analysis on the GeneGo Metacore platform demonstrated that genes coexpressed with the six PSMC genes participated in biological processes related to cancer progression. MetaCore can be used to construct downstream networks associated with biological processes from uploaded genes. By uploading PSMC coexpressed genes from the METABRIC database into the Metacore platform, we found that several cancer progression-related pathways were correlated with genes of the PSMC family (Supplementary Figures 4, 5 and Supplementary Table 2), including "Cytoskeleton remodeling_Regulation of actin cytoskeleton organization by the kinase effectors of Rho GTPases", "Cell cycle_Role of APC in cell cycle regulation", "Cell cycle_Chromosome condensation in prometaphase", "Cell cycle_Nucleocytoplasmic transport of CDK/Cyclins", and "Transcription_Role of heterochromatin protein 1 family in transcriptional silencing" (Figure 7). Next, the STRING platform was used to externally validate and search for potential protein-protein interactions (PPIs). The resulting network with a core cluster contained all of the genes associated with cancer progression and metastasis (Figure 8).
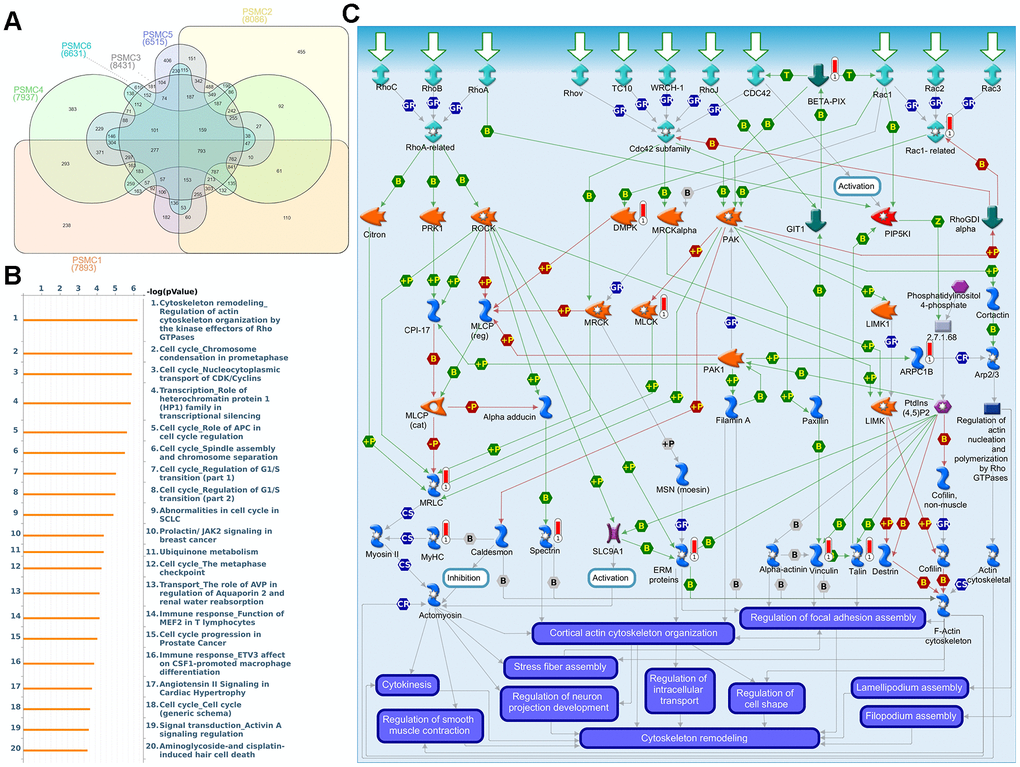
Figure 7. Coexpression of proteasome 26S subunit, ATPase (PSMC) genes and signal transduction pathways in breast cancer tissues. (A) Venn diagram of PSMC family coexpression networks in METABRIC breast cancer databases. PSMC genes were analyzed using METABRIC databases, and the intersection of coexpressed genes was plotted. (B) To explore potential networks regulated by PSMC family genes, we exported coexpressed genes and further uploaded them to the MetaCore platform for a pathway analysis. (C) The MetaCore pathway analysis of "biological processes" indicated that “Cytoskeleton remodeling_Regulation of actin cytoskeleton organization by the kinase effectors of Rho GTPases"-related pathways were correlated with breast cancer development.
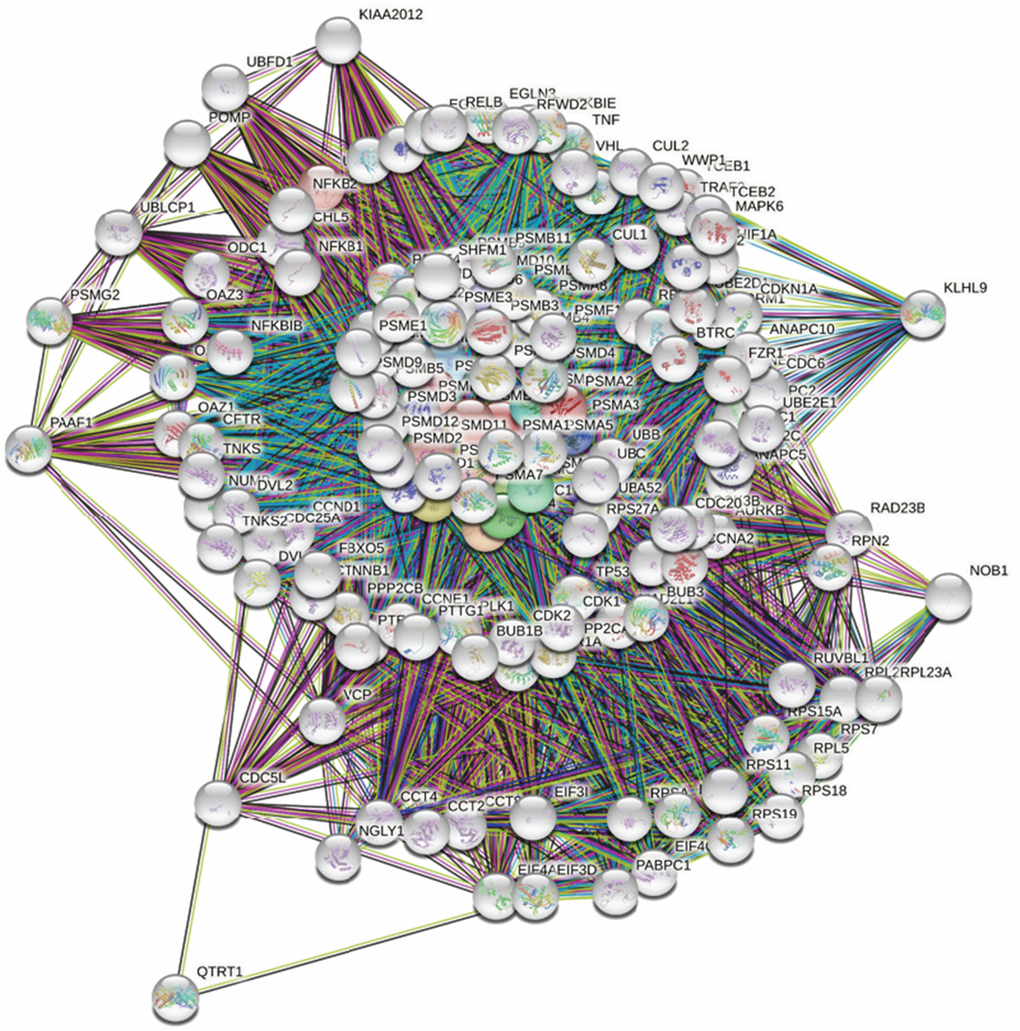
Figure 8. Network analysis of protein-protein interactions (PPIs) by the STRING platform. Genes associated with the proteasome 26S subunit, ATPase (PSMC) family were uploaded to the STRING platform to establish the network. Using k-means clustering, the network was further separated into different clusters.
Next, we explored whether individual genes of the PSMC family regulate specific pathways and networks in breast cancer development. We obtained coexpression profiles for PSMC1 from TCGA and METABRIC breast cancer datasets. Afterward, GeneGo Metacore annotations of each biological process suggested that genes coexpressed with PSMC1 were involved inG-protein-coupled receptor (GPCR)- and apoptosis-related pathways and networks such as “Chemotaxis_Lysophosphatidic acid signaling via GPCRs”, “Development_Positive regulation of WNT/Beta-catenin signaling in the cytoplasm”, and “Apoptosis and survival_Regulation of apoptosis by mitochondrial proteins”, and play essential roles in breast cancer (Supplementary Figure 6 and Supplementary Table 3). PSMC2-related genes were involved in Wnt- and hypoxia-related pathways and networks such as “Development_Negative regulation of WNT/Beta-catenin signaling in the cytoplasm” and “Transcription_HIF-1 targets”, which may be involved in breast cancer (Supplementary Figure 7 and Supplementary Table 4). Genes coexpressed with PSMC3 participated in processes of cytoskeleton- and organization-related pathways and networks such as “Cytoskeleton remodeling_Regulation of actin cytoskeleton organization by the kinase effectors of Rho GTPases” (Supplementary Figure 8 and Supplementary Table 5).
PSMC4-related genes were involved in mitogen-activated protein kinase (MAPK)- and inflammation-related pathways and networks such as “Signal transduction_CXCR4 signaling via MAPKs cascades” and “Signal transduction_Angiotensin II/AGTR1 signaling via Notch, Beta-catenin and NF-κB pathways”, which may participate in breast cancer (Supplementary Figure 9 and Supplementary Table 6). Genes found to be coexpressed with PSMC5 were involved in oxidative stress- and cell adhesion-related pathways and networks such as “Oxidative stress_ROS-induced cellular signaling” and “Cell adhesion_Tight junctions” (Supplementary Figure 10 and Supplementary Table 7). PSMC6-coexpressed genes were involved in calcium- and hormone-related pathways and networks such as “Signal transduction_Calcium-mediated signaling” and “Reproduction_Gonadotropin-releasing hormone (GnRH) signaling ”, which could participate in breast cancer (Supplementary Figure 11 and Supplementary Table 8).