Processing of single-cell RNA-seq profiles and screening of MSC-associated marker genes
The flowchart demonstrates the overall design and process about this study (Figure 1). The variance analysis revealed the top 10 significantly DEGs across the cell samples (Supplementary Figure 1A). The principal component analysis (PCA) method screened the significantly correlated genes in each component. The top 30 significantly correlated genes were shown via heatmap and dot plot in Supplementary Figure 1A, 1B. The first 35 PCs represented the main differences between the cells (Supplementary Figure 1B). According to the UMAP algorithm and marker gene annotation, 2011 MSCs were identified in metastatic tissue and primary tissue (Figure 2A). A total of 7012 DEGs between metastatic tissue and primary tissue of MSCs were identified. We downloaded the OS bulk data of GSE33382 and conducted differential expression analysis. The results showed that MT1G was highly expressed in tumor tissues compared with normal tissues (Figure 2B). Furthermore, MT1G was more highly expressed in metastatic tumor tissues than in primary tumor tissues (Figure 2C). We validated MT1G expression using 13 OS tissues and 13 adjacent cancerous tissues as the validation dataset. MT1G expression levels were confirmed using IHC (Figure 1D, 1E).
Figure 1. Research flowchart. The research process of this study is depicted in Figure 1.
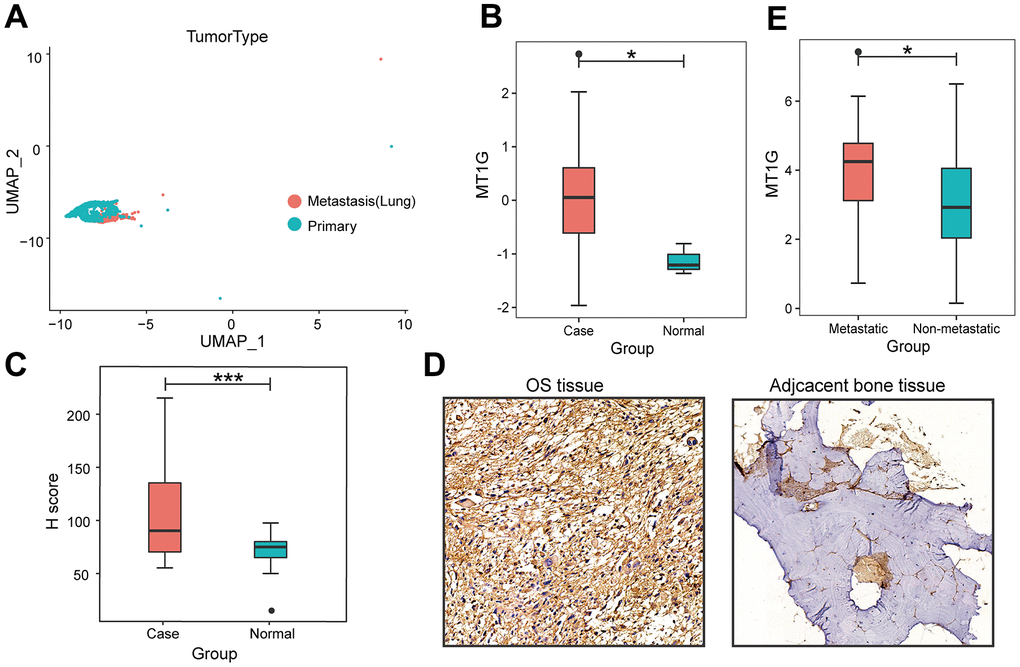
Figure 2. Characterization of single-cell RNA-seq profiles. (A) The clustering result of 2011 MSCs using the UMAP nonlinear dimensional reduction method colored by tissue origin. (B) MT1G was obviously more highly expressed in tumor tissues than in normal tissues. (C) In contrast to the primary tumor tissues, MT1G was obviously more highly expressed in metastatic tumor tissues. (D) Quantification of MT1G IHC staining in OS tissues (n=13) and adjacent cancerous tissues (n=13). (E) High/low H score of MT1G ICH images. (ns, p >0.05; *p <0.05; **p <0.01; ***p <0.001).
Identification of DEGs
After screening for DEGs between high and low MT1G-expression groups (Figure 3A), we found 134 upregulated genes and 103 downregulated genes from the high and low MT1G expression groups (Supplementary Table 1), for a total of 237 coexpressed genes (Figure 3C). The TARGET cohort was subsequently divided into high/low MT1G expression groups using the same strategy (Figure 3B), and 160 upregulated genes and 557 downregulated genes were screened (Supplementary Table 2 and Figure 3D). Finally, we screened 12 common DEGs (8 upregulated and 4 downregulated) from the two DEG groups (Figure 3E, 3F).
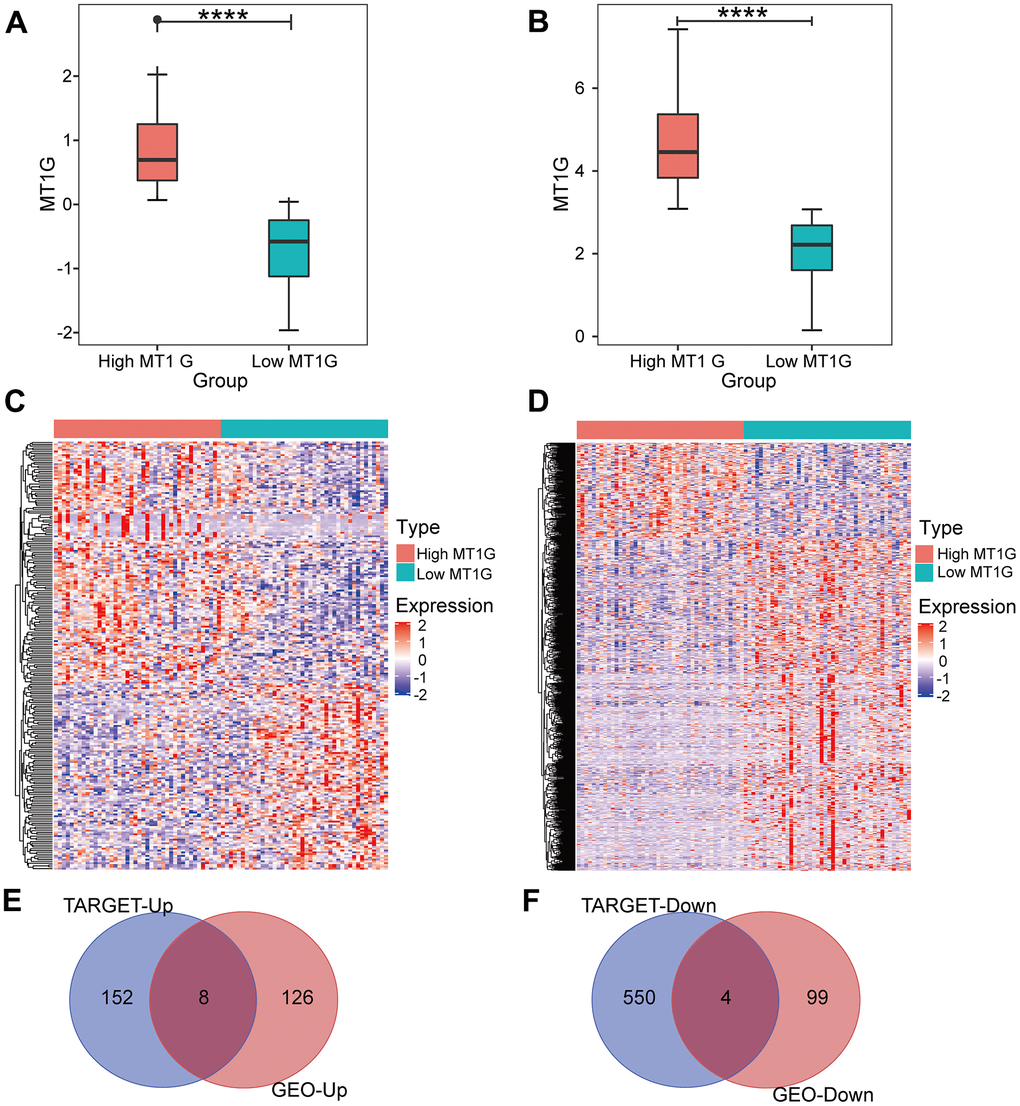
Figure 3. Expression analysis for MT1G and coexpressed genes. (A) MT1G expression in the GSE33382 cohort. (B) MT1G expression in the TARGET cohort. (C) The median expression level of MT1G in OS samples in the GSE33382 data set was used to divide patients into high/low expression groups, and the significant DEGs between the two groups were displayed in the form of a heatmap. (D) The median MT1G expression value of OS samples in the TARGET data set was used to sort the patients into high/low expression groups, and the significant DEGs between the two groups were displayed in the form of a heatmap. (E) Venn diagram of the intersection of upregulated DEGs in the GEO and TARGET datasets. (F) Venn diagram of the intersection of downregulated DEGs in the GEO and TARGET datasets. (ns, p >0.05; *p <0.05; **p <0.01; ***p <0.001).
Functional enrichment analysis and GSEA
KEGG and GO enrichment results showed that the core genes were mainly enriched in the following pathways: the p53 signaling pathway, human cytomegalovirus infection, tumor-associated pathways, detoxification of copper ions, and dendritic shaft and neuroligin family protein binding (Figure 4 and Supplementary Tables 3–6). The enriched terms for the TARGET cohort were the PI3K-Akt signaling pathway, Rap1 signaling pathway, copper ion detoxification, intrinsic components of the plasma membrane and phosphatase binding. (Figure 5 and Supplementary Tables 7–10). We selected c2.cp.kegg.v7.4.entrez as the reference gene set for our GSEA. In the GEO cohort, DEGs mainly involved spliceosome and ECM receptor interactions (Figure 6A). In the TARGET cohort, DEGs were involved in spliceosome, ribosome and cytokine-cytokine receptor interactions (Figure 6B).
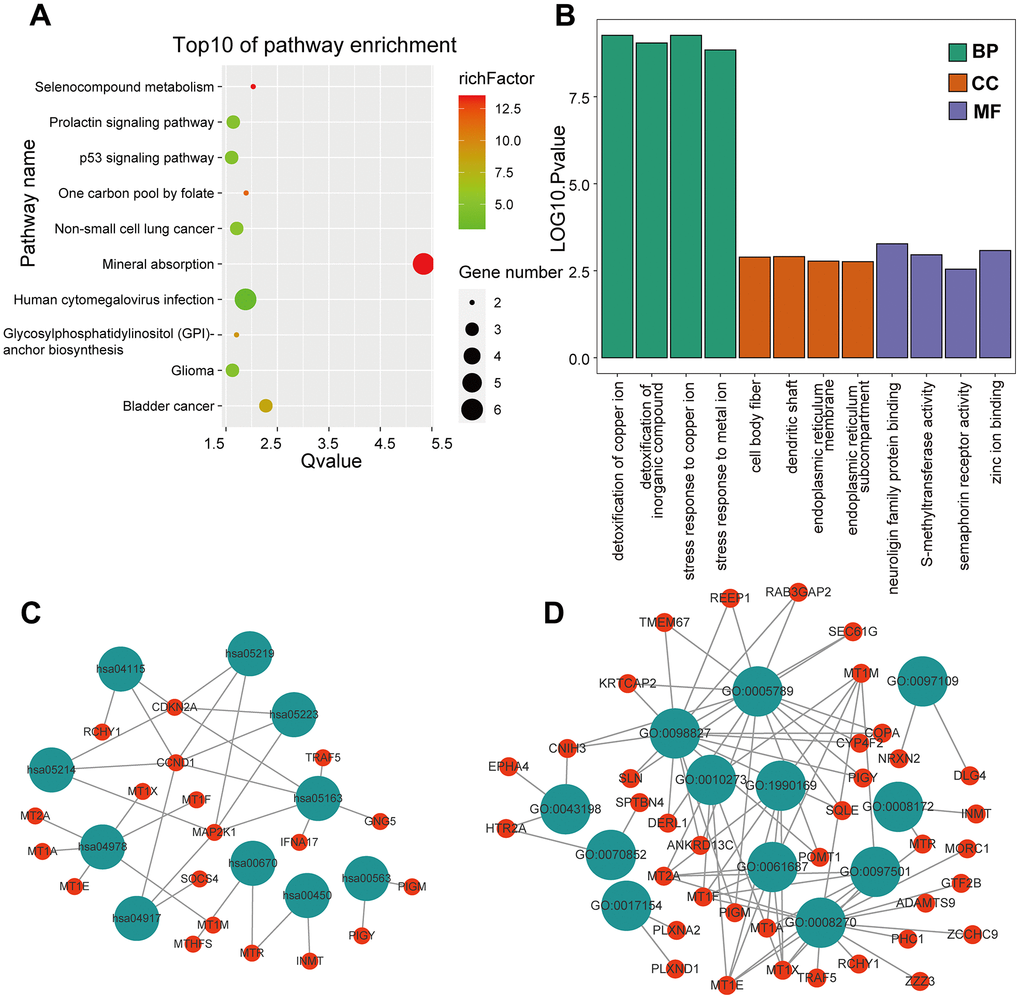
Figure 4. GO and KEGG enrichment analysis in the GEO cohort. (A) The bubble graph package was applied to visualize the results of KEGG enrichment analysis; the bubble size represented the number of enriched genes, and the color represented the enrichment ratio in the GSE33382 data set. (B) The bar graph for GO enrichment analysis; length represented significance in the GSE33382 data set. (C) A functional enrichment network based on KEGG analysis of the GSE33382 data set. (D) A functional enrichment network based on GO analysis of the GSE33382 data set.
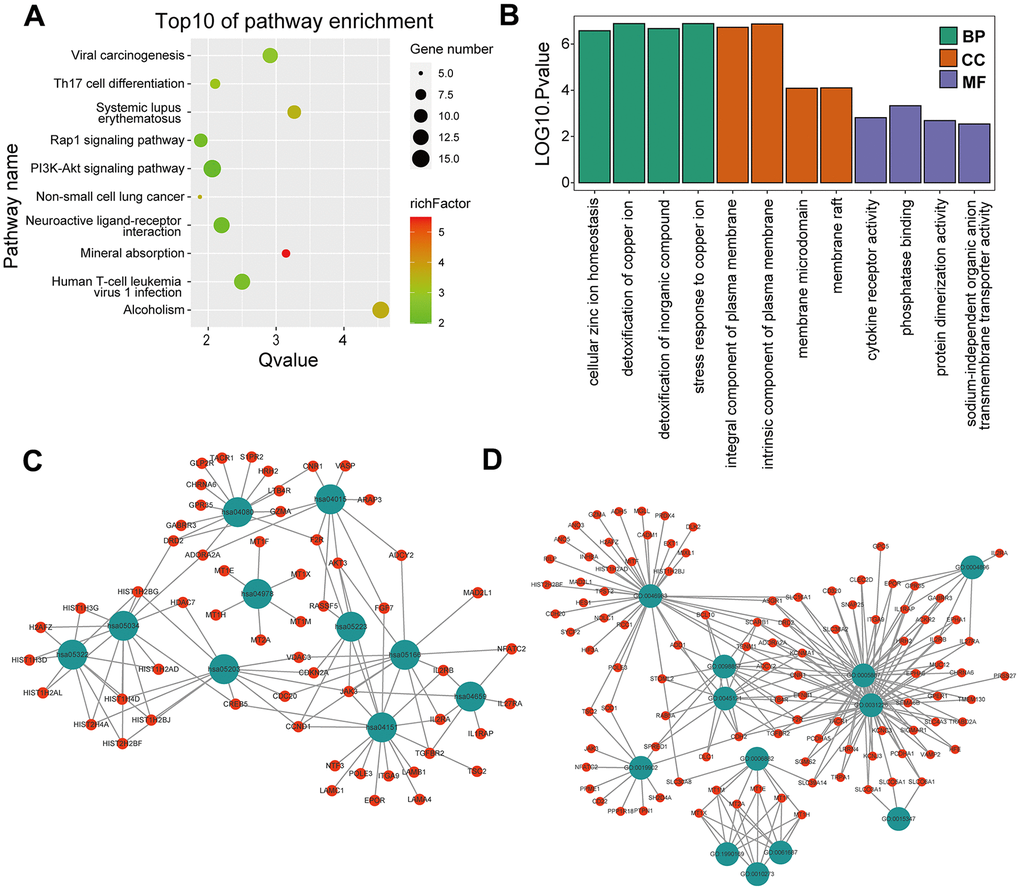
Figure 5. GO and KEGG enrichment analyses in the TARGET cohort. (A) The bubble graph package was applied to visualize the results of KEGG enrichment analysis; the bubble size represented the number of enriched genes, and the color represented the enrichment ratio in the TARGET data set. (B) The bar graph for GO enrichment analysis; length represented significance in the TARGET data set. (C) A functional enrichment network based on KEGG analysis of the TARGET data set. (D) A functional enrichment network based on GO analysis of the TARGET data set.
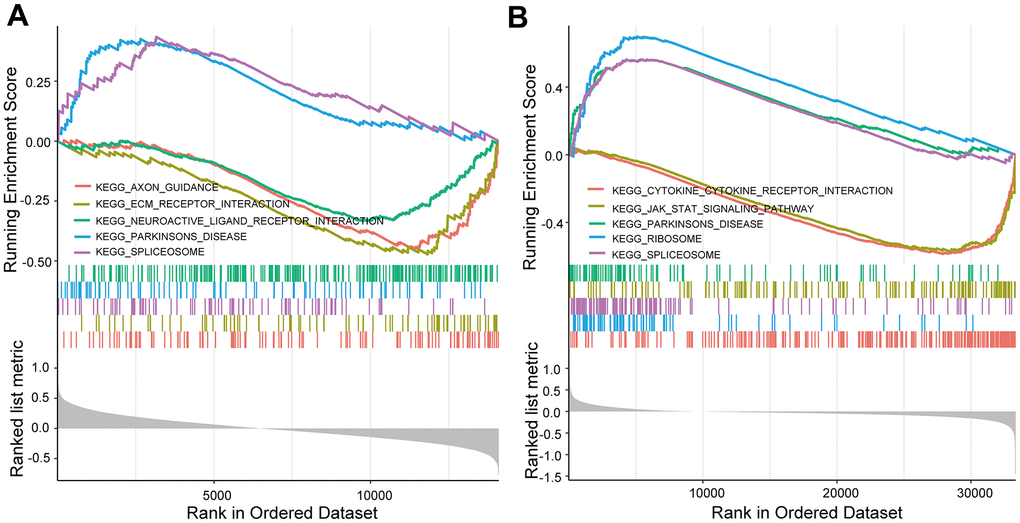
Figure 6. GSEA enrichment analysis. (A) GSEA enrichment analysis in the GEO cohort. (B) GSEA enrichment analysis in the TARGET cohort.
Construction of PPI networks and hub-gene screening
We constructed PPI networks from the GEO and TARGET cohort DEGs separately using the STRING tool (Figure 7A, 7C) and used the MCODE clustering algorithm to filter hub gene clusters. For the GEO cohort, screened hub genes were linked in the following networks: MCODE_1 (CCND1, MAP2K1, CDKN2A, MCL1, CFLAR), MCODE_2 (SYN1, PPFIA2, NRXN2), MCODE_3 (MRPL35, MRPS18C, MRPL40), MCODE_4 (MAGEA3, XAGE1A, SSX5), MCODE_5 (TMEM147, KRTCAP2, SEC61G), MCODE_6 (EPHA4, PLXND1, PLXNA2) and MCODE_7 (ILF2, CHTOP, MAGOH; Figure 7B). Hub genes in the TARGET cohort were involved in the following networks: MCODE_1 (BIRC5, CDCA8, CDKN3, CKS2, H2AFZ, HDAC7, HIST1H2AD, HIST1H2AL, HIST1H2BG, HIST1H2BJ, HIST1H3D, HIST1H3G, HIST1H4D, HIST2H2BF, HIST2H4A, KDM1A, MAD2L1, NDC1, PBK), MCODE_2 (MT1E, MT1F, MT1G, MT1H, MT1M, MT1X, MT2A, PTPN1), MCODE_3 (DDX23, LSM7, MAK16, MRTO4, NOLC1, POLR1E, PRPF3, PRPF38A, RRP1, SNRNP40), MCODE_4 (HYOU1, SEC61A1, SRPR, SRPRB), MCODE_5 (CCND1, CDC20, WDR77), MCODE_6 (LAMA4, LAMB1, LAMC1), and MCODE_7 (ADCY2, ADORA2A, AK2, CNR1, DRD2, PDE3A, PDE4B; Figure 7D).
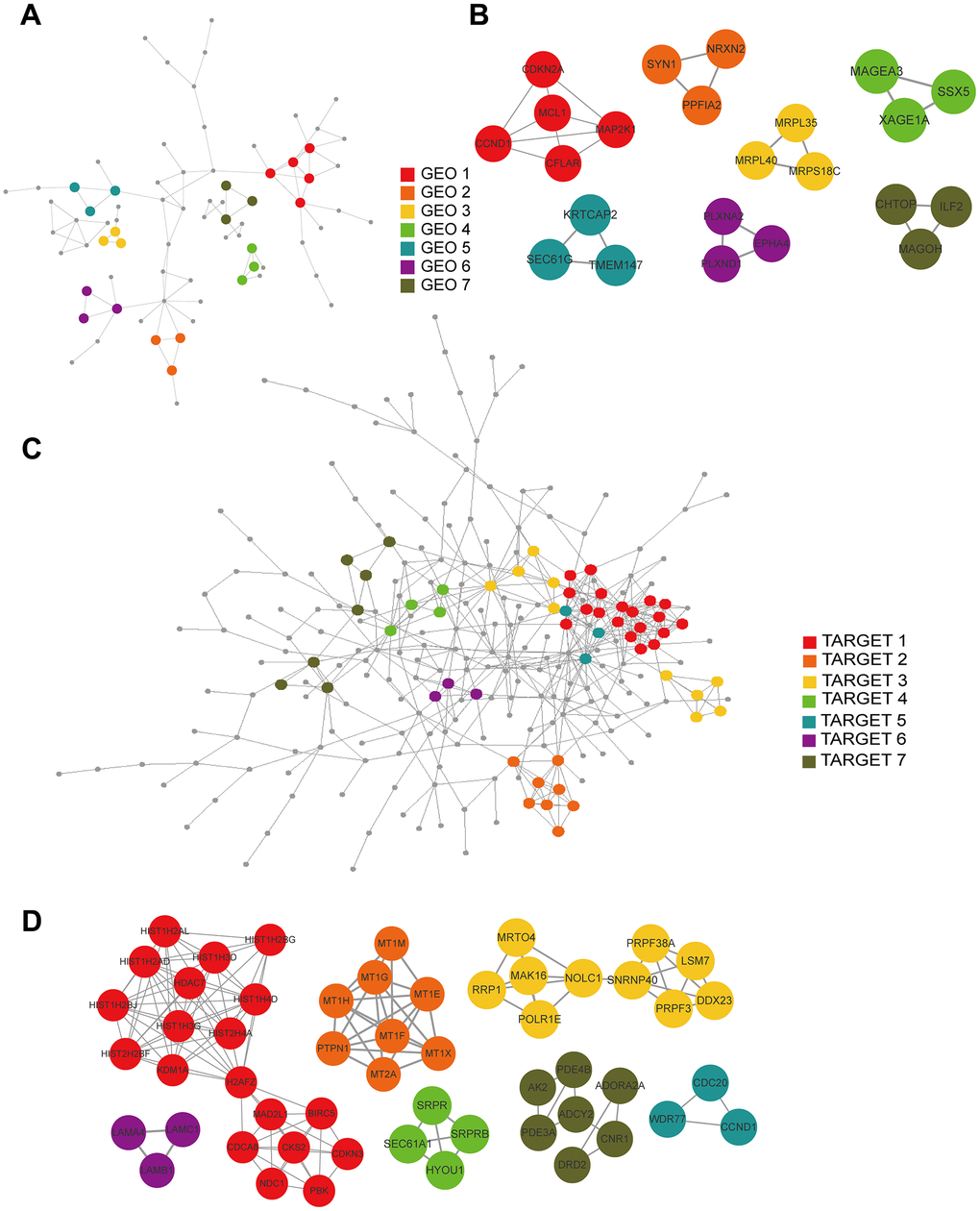
Figure 7. PPI and hub gene cluster network construction. (A) A PPI network from the GSE33382 data set. (B) Seven hub gene clusters obtained by the MCODE clustering algorithm from the GSE33382 data set. (C) A PPI network from the TARGET data set. (D) Seven hub gene clusters obtained by the MCODE clustering algorithm from the TARGET data set.