Characterization
of Geneswitch drivers in adult flies using the UAS-GFP reporter
To
facilitate the screen of apoptosis and senescence-regulatory genes for life
span effects, several Geneswitch system drivers were characterized for their
tissue-specificity of transgene activation using a UAS-GFP reporter, both in
adult flies and during larval development. The UAS-GFP reporter employed was
"UAS-ultraGFP" which contains multiple copies of a UAS-eGFP construct, and
yields particularly high levels of GFP expression [18]. Three Geneswitch
system drivers were characterized: The Act-GS-255B driver strain contains
multiple inserts of a construct in which the promoter from the cytoplasmic
actin gene Actin 5C drives Geneswitch, and is expected to yield
tissue-general expression [16]. The Elav-GS driver contains Geneswitch under
control of the Elav gene promoter and produces nervous-system-specific
expression [19]. Finally the whole-body fat-body Geneswitch driver strain
("WB-FB-GS") contains both a head fat-body driver (S1-32) and a
body-fat-body driver (S1-106) [20-22], and is expected to yield expression
in the fat-body tissue throughout the animal. The three driver strains were
crossed to the UAS-ultraGFP reporter strain to produce adult progeny containing
both the driver and reporter constructs, and the flies were cultured in the
presence and absence of drug for two weeks. GFP expression was scored in live
adult flies as well as in several dissected tissues (Figure 1). The
Act-GS-255B driver was found to yield tissue-general expression of the
UAS-ultraGFP reporter in adult flies. In whole adults, GFP expression was
observed throughout the body of both males and females, with greater expression
levels observed in females relative to males. Similarly with heads dissected
in half and bodies dissected in half, expression was observed in all tissues,
including abundant expression in nervous system, muscle (including flight
muscle), and fat-body tissue. Note that flight muscle in male has lower
expression than flight muscle in female, however inspection of the GFP-only
image for male flight muscle (inset) reveals expression throughout this
tissue. Abundant expression was also observed throughout dissected gut tissue,
ovary and testes. The expression level was greater in some regions of the gut
than others, however all regions of the gut exhibited staining, as revealed by
inspection of the GFP-only images (inset).
All tissues observed showed significant GFP expression, and therefore we
conclude that Act-GS-255B yields truly tissue-general expression in adult flies. The WB-FB-GS driver produced GFP expression in the
head-fat-body and body-fat-body tissues,
as expected, as well as in the gut and testes, and very faint expression in
ovary; there was no detectable expression in nervous, muscle, or other
tissues. Notably, the expression in adult male head fat body was much reduced
relative to female head fat body, consistent with recent characterization of
the fat body drivers using a LacZ reporter [17]. Finally, the Elav-GS driver produced abundant expression in the brain and ventral nerve cord, as expected, and there
was no detectable expression in any other tissues; for example, the muscle, gut
and gonads were clearly negative. Note the GFP-only image for the gut (inset)
shows a lack of expression. The Elav-GS driver was found to produce similar
levels of UAS-GFP reporter expression in male versus female in our experiments.
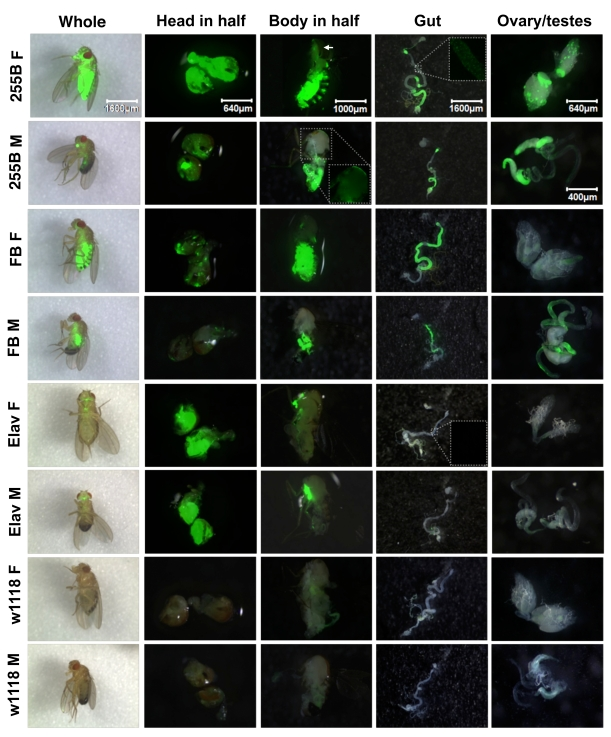
Figure 1. Expression pattern produced by Geneswitch drivers and UAS-GFP reporter in adult flies. The indicated GeneSwitch
drivers Act-GS-255B ("255B"), Elav-GS ("Elav") and WB-FB-GS ("FB") were
crossed to the UAS-ultraGFP reporter and adult progeny containing both
constructs were scored for GFP expression in various tissues. Control
flies were generated by crossing UAS-ultraGFP to white1118strain flies to produce progeny containing only UAS-ultraGFP. Age-synchronized
flies were cultured in the presence and absence
of the drug RU486 for two weeks prior to assay, and GFP expression was
scored in whole adult flies and dissected tissues, as indicated. Each
image is the overlay of the visible light and GFP images. Insets show
details of the regions boxed in white, GFP image only. M = male, F =
female. Pictures were taken at the magnification of 20X, 50X, 32X, 20X,
50X, and 80X, for whole fly, head in half, body in half, gut, ovary, and
testes, respectively. The white arrow indicates a region of 255B Female
flight muscle that is obscured by a fragment of cuticle.
Characterization
of Geneswitch drivers in larvae using the UAS-GFP reporter
The Geneswitch driver strains were also
scored for expression patterns in 3rd instar larvae and dissected
tissues (Figure 2). The Act-GS-255B driver was found to
yield tissue-general expression, including abundant expression throughout the
body of whole 3rd instar larvae, as well as in dissected brain, gut,
salivary gland, imaginal discs and fat-body tissues; all tissues observed
showed abundant GFP expression (Figure 2A). The inset for the Act-GS-255B 3rd
instar larval brain shows detail of the
GFP-only image, and indicates that expression was present throughout the brain,
with higher-level expression in a subset of cells. The WB-FB-GS driver was
found to drive abundant expression in salivary gland and anterior midgut, but
notably no expression in any other larval tissues including larval fat-body.
Finally the Elav-GS driver produced abundant expression in larval nervous
system and no detectable expression in any other larval tissues. The inset for
the Elav-GS 3rd instar larval brain shows detail of the GFP-only image, and shows that expression was present throughout the
brain, with higher-level expression in a subset of cells. Notably this subset
of cells was different from that observed above with Act-GS-255B. Each of the
three drivers was found to produce similar patterns of expression in 1st
and 2nd instar larvae as well (Figure 1B). When the Act-GS-255B
driver was induced using dilutions of RU486 drug in the culture media, it
produced a dose-response of GFP expression in 3rd instar larvae
(Figure 1D), as well as in adult flies (data not shown).
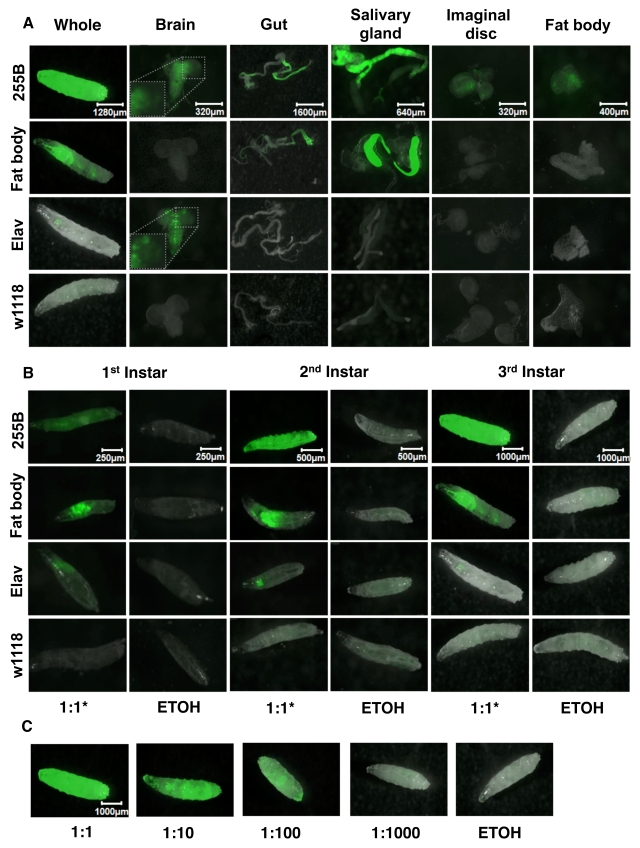
Figure 2. Expression pattern produced by GeneSwitch drivers and UAS-GFP reporter in larvae. The crosses are the same as
Figure 1, but larvae were cultured in the presence and absence of drug in
the food, from hatching to the indicated developmental stage. A.
Expression patterns in 3rd instar larvae and dissected tissues.
For the Elav-GS driver ("Elav") a 1:10 dilution of drug was used because of
the toxic effects of drug observed in larvae with this driver. Pictures
were taken at the magnification of 25X, 100X, 20X, 50X, 100X, 80X, for
whole larvae, brain, gut, salivary gland, imaginal discs, and fat body,
respectively. B. Expression patterns in the three larval stages. For Elav-GS
a 1:10 dilution of drug was used to avoid toxic effects. GFP pictures were
taken at the magnification of 100X, 50X, 25X, for 1st instar, 2nd
instar, and 3rd instar, respectively. C. Expression in 3rd
instar larvae using Act-GS-255B and titrations of drug. ETOH indicates the
ethanol solvent for the drug alone. Pictures were taken at the
magnification of 25X.
Effect
of apoptosis and senescence-regulatory gene over-expression on life span
Fourteen apoptosis and senescence
regulatory genes were chosen for analysis based on their relevance to human
apoptosis and senescence pathways and the availability of reagents for Drosophila. Ras85D is a Drosophila homolog of the human oncogene Ras
that encodes a GTPase involved in signal transduction.
Ras85D activated form contains an amino acid substitution that causes Ras
to be constitutively active [23], and Ras85D dominant negative (DN)
form contains an amino acid substitution that causes it to inhibit the
endogenous Ras protein [23,24]. Wingless is a Drosophila homolog of
the human Wnt signaling protein involved in development and tumorigenesis
[25]. Pk61C is a serine/threonine protein kinase related to human PDK-1 and
involved in growth signaling [26]. DIAP1 is a Drosophila member of the
inhibitor of apoptosis protein (IAP) family [27]. Baculovirus p35 is a
caspase inhibitor protein also related to the IAPs. Nemo (nmo)
is the Drosophila homolog of a human protein kinase regulatory subunit
involved in NF-kappaB signaling pathway [28]. Egfr is the Drosophila
homolog of the human epidermal growth factor receptor [29]. The Drosophilapointed (pnt) gene encodes a transcription factor homologous to
human Ets1 that is involved in the Ras signaling pathway. The DrosophilaMatrix metalloproteinase 2 gene (Mmp2) is involved in tissue
remodeling and tumor progression and is related to a family of human matrix
metalloproteinases [30]. The DrosophilaStat92E
gene encodes a homolog of the human Stat transcription factor, which is a
target of the Jak-Stat growth-regulatory pathway [31]. The Drosophilapuckered
(puc) gene encodes a phosphatase homologous to the human VH-1 family
that antagonizes JNK signaling, and heterozygous puc mutant flies have
been reported to have increased stress resistance and life span [32,33]. The DrosophilaSphingosine kinase 2 (Sk2) gene encodes
a lipid kinase involved in activation of protein kinase C-family signaling, and
the human homolog Sphk2 is implicated in regulation of apoptosis [34].
Finally the CG14544 gene encodes a predicted methyltransferase, and the Drosophilabantam (ban) gene encodes a micro-RNA that inhibits expression of
pro-apoptotic genes [35]. Each of these genes of interest was over-expressed in
adult flies or during larval development, and assayed for effects on adult fly
life span.
To control for any possible effects of
the Geneswitch system and the RU486 drug itself, life span was assayed in flies
that were the progeny of Act-GS-2555B driver crossed to either Oregon-R (Or-R)
wild-type strain or to the w1118 control strain, to produce
progeny containing only the driver. In these control flies, treatment with
drug produced small, but statistically significant reductions in life span in
both male and female adults: treatment during adulthood reduced mean life span
by -4% to -10%, while treatment in larval stages reduced adult life span by -8%
to -16% (Figure 3A, B; Figure 4A, B; Tables 2, 3). There were no significant
increases in life span in control flies treated with RU486 in any of the
replicate experiments. These data indicate that in these experiments, when the
Act-GS-255B driver is present, the RU486 can cause small but significant
reductions in adult life span, and this effect must be taken into account when
interpreting the effects of transgene over-expression. Other studies [22],
including ones from our own laboratory using the Act-GS-255B driver [36], found
no negative effects of RU486 on adult fly life span. We conclude that the small
negative effects observed here result from differences in the lot of RU486
drug, and/or small differences in effective concentrations due to specifics of
media preparation. To confirm that the Act-GS-255B driver can produce increased
life span, it was used to drive over-expression of the dominant p53
allele (p53-259H). Over-expression of p53-259H in adult flies
using the ubiquitous Act-GS-255B driver produced increased median life span in
females (+8%) but not males (-2.8%), and no life span increase when expressed
in larvae (Table 3). These results are consistent with previous studies showing
that expression of p53-259H in the adult nervous system with the Elav-GS
driver can cause increased life span in females [14], and confirms that the
Act-GS-255B driver can indeed produce increased life span when combined with an
appropriate target gene.
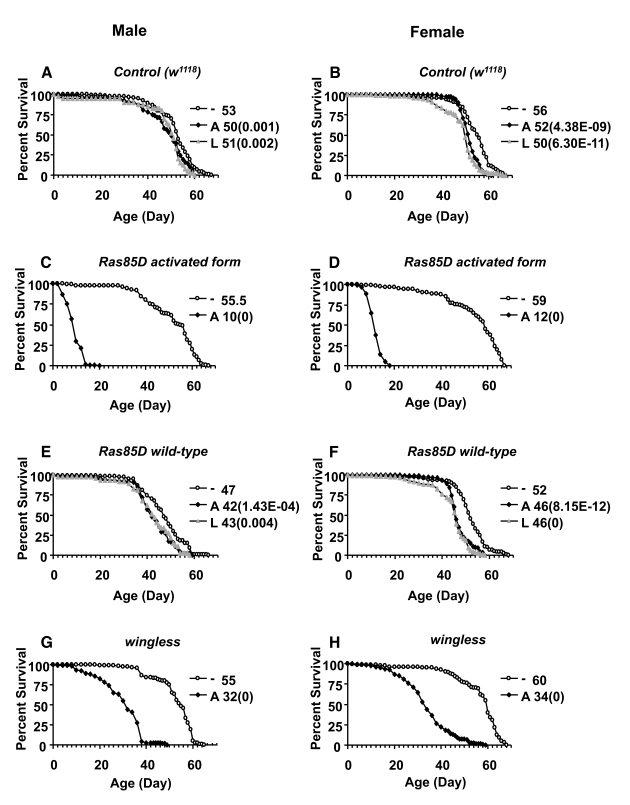
Figure 3. Effect of transgene over-expression on survival of adult flies. Apoptosis and
senescence-related genes wingless, Ras85D, and Ras85D
activated form were over-expressed during larval development or in
adults, and assayed for effects on adult life span in male and female
flies, as indicated. The life span assays were performed at 29°C. Open circles represent the no-drug control
("-"). Solid squares represent adults treated with drug ("A"). Grey
triangles represent larvae on drug ("L"). Survival curves are plotted as a
function of adult age in days. Median life span of each cohort is presented
along with p value for log rank test (in parentheses). (A, C,
E, G) male flies. (B, D, F, H) female flies. (A, B) Control
flies containing the driver and no target transgene. (C, D) Ras85D
activated form. (E, F) Ras85D wild-type. (G, H) wingless.
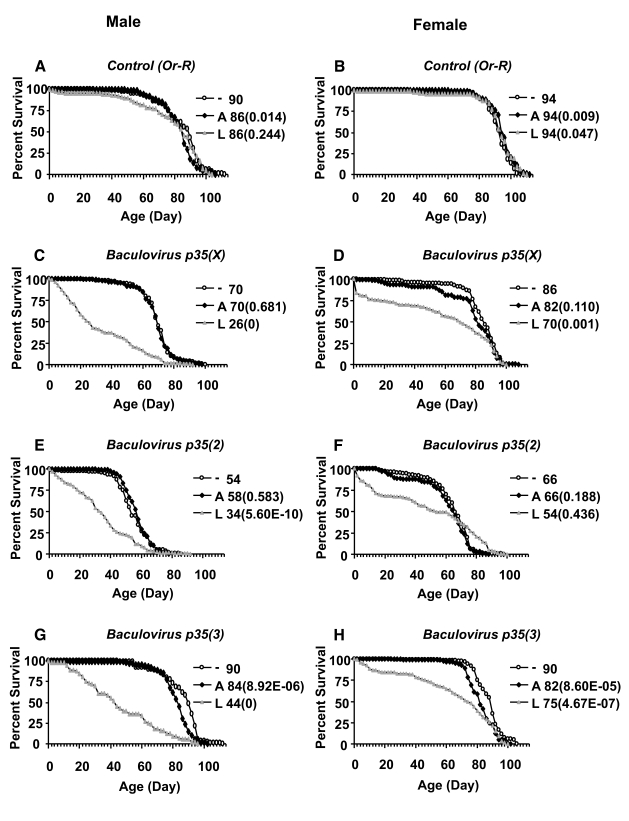
Figure 4. Effect of Baculovirus p35 over-expression on survival of adult flies. Baculovirus p35
transgenes inserted on the X chromosome, chromosome 2, and chromosome 3
were over-expressed during larval development or adult stage, as indicated.
The life span assays were performed at 25°C. Open circles represent the no-drug control ("-"). Solid squares
represent adults treated with drug ("A"). Grey triangles represent larvae
on drug ("L"). Survival curves are plotted as a function of adult age in
days. Median life span of each cohort is presented along with p
value for log rank tests (in parentheses). (A, C, E, G) male
flies. (B, D, F, H) female flies. (A,B) Control flies
containing the driver and no target transgene. (C, D) Baculovirus
p35 transgene on X chromosome. (E, F) Baculovirus p35 transgene
on second chromosome. (G, H) Baculovirus p35 transgene on
third chromosome.
Most
of the genes tested by over-expression with the ubiquitous Act-GS-255B driver
did not affect life span to an extent greater than the small changes observed
with the control flies. However, Ras activated form transgene was
lethal when expressed in larvae, and reduced both male and female life span by
-80% when expressed in adults (Figure 3C, D; Table 2). Over-expression of
wild-type Ras or a Ras dominant-negative allele was not lethal to
larvae, and produced only small decreases (-4% to -12%) in both male and female
adult life span (Figure 2 E, F; Table 2), thereby in the range of negative
effects observed with control flies. Over-expression of the wingless
gene was found to be lethal to male and female larvae, using two independent wingless
transgenes (Table 2). Over-expression of wingless in adult flies
produced significant reductions in both male and female life span: ~-42% with
one wingless transgene (Figure 3 G, H) and ~-10% with the other
transgene (Table 2).
Finally,
the tissue-general Act-GS-255B driver was used to over-express three different
transgenes encoding the caspase inhibitor Baculovirus p35, during larval
development and in adult flies (Figure 4; Table 3). Over-expression of Baculovirusp35 in adult flies using the tissue-general Act-GS-255B driver produced
only small decreases in life span that were within the range observed with
control flies, suggesting there were no significant effects in adults. In
contrast, when Baculovirus p35 was over-expressed during larval
development using the tissue-general driver, it reduced the mean life span of
male and female adults by -20% to -50%. Interestingly, over-expression of each
of the three independent Baculovirus p35 transgenes during larval development
produced an unusual biphasic-shaped survival curve in adult females (Figure 4
D, F, H), suggesting the presence of a subset of adult female flies with unchanged
or even increased life span. A Gompertz-Makeham
model was found to give the best fit to the life span data for females in which Baculovirus p35 was over-expressed during larval development (Figure 5;
Table 4). This analysis revealed that the decrease in mean life span was due
to increased age-independent mortality. When the age-independent mortality was
removed and the data re-plotted, it revealed a subset of female flies with
unchanged (Figure 5 B, F) or increased life span (Figure 5D).
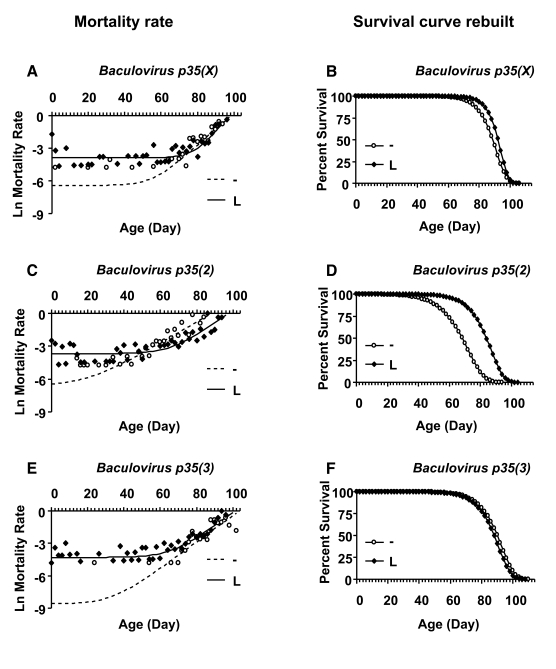
Figure 5. Mortality rate analysis of female larvae with and without Baculovirus p35 transgene expression. Open circles represent the
no-drug control ("-"). Solid squares represent larvae cultured with drug
("L"). (A, B) Baculovirus p35 transgene on X chromosome. (C,
D) Baculovirusp35 transgeneon second chromosome. (E, F) Baculovirus
p35 transgene on third chromosome. (A, C, E) Plots of
natural-log mortality rate vs. age in days. (B, D, F) The data
were fitted to the Gompertz-Makeham model, which best described the
mortality rate. The age-independent mortality was removed and the survival
curves were re-drawn using only the Gompertz components. Mortality rate
analysis showed that age-independent mortality was significantly higher for
female larvae on drug versus control for all three Baculovirus p35
lines (Table 4).
Two independent Baculovirus p35
transgenes were also over-expressed in adult flies using the head-fat-body
driver S1-32, and the whole-body fat-body driver (S1-32
plus S1-106), and during larval development using the whole-body
fat-body driver, however no consistent effects on life span were observed
(Table 3).
The
nervous system-specific Elav-GS driver was also used to over-express two baculovirus
p35 transgenes. In adults the Elav-GS driver itself had little to no
effect on life span, and over-expression of baculovirus p35 in adults
using Elav-GS had no consistent effects on life span (Table 3). In contrast,
when drug was administered to larvae, the Elav-GS driver itself was associated
with significant decreases in life span in both males (~-30% to -40%) and
females (~-25%), and significantly reduced the number of male adults, and no
effects of the baculovirus p35 transgenes on life span could be
identified in this background (Table 3). In an attempt to reduce this
background toxicity and allow assay of baculovirus p35 transgenes with
the Elav-GS driver in larvae, a 1:10 dilution of drug was used. Under these
conditions the life span reductions caused by drug in males and females were
smaller (~-2% to -12%), and the number of males obtained was approximately
normal, however no increases in life span were observed upon over-expression of baculovirus p35 (Table 3).
The
muscle-specific MHC-GS driver was used to drive over-expression of several
transgenes in adult flies, however the MHC-GS driver itself was found to cause
a significant RU486-dependent decrease in life span in both males and females
(~-20% to -30%), and none of the target transgenes tested produced a
significant life span increase in this background (Table 3).
Table 1. Starting Stocks
| St# | Genotype | Notes | Abbreviation |
| 1 | w; GS-Actin255-B:+ | Ubiquitous
GeneSwitch 255B Driver | 255B |
| 2 | w; GS-Actin255-A;+ | Ubiquitous
GeneSwitch 255A Driver | 255A |
| 3 | w; P{Switch}bun[Switch
1-32];+ | GeneSwitch
Head Fat Body Driver | S32 |
| 4 | w; P{Switch}S1-106
P{Switch}bun[Switch 1-32];+ | GeneSwitch
Head & Thorax-Abdomen Fat Body Driver | S106 S32 |
| 5 | yw; +; GS-Elav | GeneSwitch
Elav Driver | Elav |
| 6 | yw; Sp/CyO,FLP.lacZ;
MHC:GS | GeneSwitch
Muscle Driver | Sp/CyO, MHC |
| 7 | Oregon R ( +; +; +) | wild type | |
| 8 | w1118; +;
+
| wild type | |
| 9 | P{UAS.p35.H}BH3,w*;+;+ | UAS-p35 on chromosome 1 | p35 |
| 10 | w*; P{UAS.p35.H}BH1;+ | UAS-p35 on chromosome 2 | p35 |
| 11 | w*; +; P{UAS.p35.H}BH2 | UAS-p35 on chromosome 3 | p35 |
| 12 | w1118; +;
P{UAS-Ras85D.V12}TL1 | UAS-Ras85D activated form | Ras act |
| 13 | w*; P{UAS-Ras85D.K}5-1;+ | UAS-Ras85D WT form | Ras WT |
| 14 | P{UAS-Ras85D.N17}TL1,
w1118; +; + | UAS-Ras85D DN form | Ras DN |
| 15 | w*; P{UAS-wg.H.T:HA1}3C;+ | UAS-wg on chromosome 2 | wga |
| 16 | w*; +;
P{UAS-wg.H.T:HA1}6C | UAS-wg on chromosome 3 | wgb |
| 17 | y1 w67c23;
+;P{EPgy2}EY04093 | EP-Pk61C | Pk61Ca |
| 18 | w; +; P{EP}Pk61CEP3644/TM6,Tb | EP-Pk61C | Pk61Cb |
| 19 | w*; +; P{UAS-DIAP1.H}3 | UAS-DIAP1 | DIAP1 |
| 20 | y1 w67c23;
P{EPgy2}EY00935 | EP-nmo | nmo |
| 21 | y1 w*; +;
P{UAS-Egfr.B}32-26-1 | UAS-Egfr | Egfr |
| 22 | y1 w67c23; +;
P{EPgy2}pntEY03254 | EP-pnt | pnt |
| 23 | y1 w67c23;
P{EPgy2}Mmp2EY08942/CyO; + | EP-Mmp2 | Mmp2 |
| 24 | y1 w67c23; +;
P{EPgy2}Stat92EEY14209/TM3, Sb1 Ser1 | EP-Stat92E | Stat |
| 25 | w*; +;
P{EPgy2}pucEY09772/TM6C | EP-puc | puc |
| 26 | y1 w67c23; +;
P{EPgy2}scramb2EY01180 | EP-Sk2 | Sk2 |
| 27 | y1 w67c23; +;
P{EPgy2}EY06207 | EP-ban | ban |
| 28 | w1118; +;
PBac{WH}CG14544f01091/TM6B, Tb1 | XP-CG14544 | CG14544 |
| 29 | w1118;
+; P{GUS-p53.259H}3.1 | UAS-p53 point mutation | p53.259H |
Table 2. Life span data of apoptosis-related
gene experiments, with means, standard deviations, medians, percent change in mean and
median, and log rank p value.
| CrossMxF | RU486 | Genotype | Sex | N | Meana | Median | %Change in Mean | %Change in Median | Log Rank p Value |
| Exp1 Life
span assay using GS255B driver at 29C |
| 8-1
| -
| w/Y; 255B/+;
+ | M
| 115
| 51.53±8.66
| 53
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
+ | M
| 120
| 47.29±11.06
| 50
| -8.23
| -5.66
| 0.001
|
| L
| w/Y; 255B/+;
+ | M
| 40
| 47.28±12.15
| 51
| -8.26
| -3.77
| 0.002
|
| -
| w/w; 255B/+;
+ | F
| 128
| 54.18±8.26
| 56
| ---------
| ---------
| ---------
|
| A
| w/w; 255B/+;
+ | F
| 120
| 51.74±3.89
| 52
| -4.5
| -7.14
| 4.38E-09
|
| L
| w/w; 255B/+;
+ | F
| 120
| 48.18±8.38
| 50
| -11.07
| -10.71
| 6.30E-11
|
| 12-1
| -
| w/Y; 255B/+;
Ras act/+ | M
| 122
| 51.11±11.58
| 55.5
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
Ras act/+ | M
| 128
| 9.45±3.42
| 10
| -81.5
| -81.98
| 0
|
| L
| w/Y; 255B/+;
Ras act/+ | M
| 0
| NA
| NA
| ---------
| ---------
| ---------
|
| -
| w/w*;
255B/+; Ras act/+ | F
| 123
| 54.19±13.26
| 59
| ---------
| ---------
| ---------
|
| A
| w/w*;
255B/+; Ras act/+ | F
| 123
| 12.11±2.8
| 12
| -77.64
| -79.66
| 0
|
| L
| w/w*;
255B/+; Ras act/+ | F
| 0
| NA
| NA
| ---------
| ---------
| ---------
|
| 13-1
| -
| w/Y; 255B/Ras
WT;+ | M
| 124
| 46.65±9.01
| 47
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/Ras
WT;+ | M
| 122
| 42.3±9.13
| 42
| -9.32
| -10.64
| 1.43E-04
|
| L
| w/Y; 255B/Ras
WT;+ | M
| 47
| 42.06±10.37
| 43
| -9.84
| -8.51
| 0.004
|
| -
| w/w*;
255B/Ras WT;+ | F
| 126
| 51.31±8.64
| 52
| ---------
| ---------
| ---------
|
| A
| w/w*;
255B/Ras WT;+ | F
| 126
| 46.84±5.17
| 46
| -8.71
| -11.54
| 8.15E-12
|
| L
| w/w*;
255B/Ras WT;+ | F
| 118
| 43.66±8.85
| 46
| -14.91
| -11.54
| 0
|
| 1-14
| -
| Ras DN, w/Y;
255B/+; + | M
| 127
| 47.89±9.88
| 50
| ---------
| ---------
| ---------
|
| A
| Ras DN, w/Y;
255B/+; + | M
| 123
| 43.64±6.87
| 44
| -8.87
| -12
| 5.78E-08
|
| L
| Ras DN, w/Y;
255B/+; + | M
| 79
| 44.76±12.49
| 48
| -6.54
| -4
| 0.14
|
| -
| Ras DN, w/w;
255B/+; + | F
| 121
| 51.65±14.25
| 57
| ---------
| ---------
| ---------
|
| A
| Ras DN, w/w;
255B/+; + | F
| 125
| 51.82±8.3
| 53
| 0.32
| -7.02
| 5.09E-04
|
| L
| Ras DN, w/w;
255B/+; + | F
| 125
| 45.39±13.16
| 49
| -12.12
| -14.04
| 1.98E-09
|
| 15-1
| -
| w/Y; 255B/wga;
+ | M
| 130
| 52.56±8.37
| 55
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/wga;
+ | M
| 122
| 29.83±9.32
| 32
| -43.25
| -41.82
| 0
|
| L
| w/Y; 255B/wga;
+ | M
| 0
| NA
| NA
| ---------
| ---------
| ---------
|
| -
| w/w*; 255B/wga;
+ | F
| 122
| 55.78±12.01
| 60
| ---------
| ---------
| ---------
|
| A
| w/w*; 255B/wga;
+ | F
| 125
| 33.61±10.55
| 34
| -39.75
| -43.33
| 0
|
| L
| w/w*; 255B/wga;
+ | F
| 0
| NA
| NA
| ---------
| ---------
| ---------
|
| 16-1
| -
| w/Y;
255B/+;wgb/+ | M
| 124
| 52.31±8.81
| 56
| ---------
| ---------
| ---------
|
| A
| w/Y;
255B/+;wgb/+ | M
| 131
| 45.02±8.04
| 47
| -13.94
| -16.07
| 0
|
| L
| w/Y;
255B/+;wgb/+ | M
| 0
| NA
| NA
| ---------
| ---------
| ---------
|
| -
| w/w*;
255B/+;wgb/+ | F
| 120
| 51.29±10.38
| 53
| ---------
| ---------
| ---------
|
| A
| w/w*;
255B/+;wgb/+ | F
| 123
| 47.55±7.23
| 49
| -7.29
| -7.55
| 3.24E-10
|
| L
| w/w*;
255B/+;wgb/+ | F
| 0
| NA
| NA
| ---------
| ---------
| ---------
|
| 17-1
| -
| w/Y; 255B/+;
Pk61Ca/+ | M
| 122
| 47.9±9.67
| 47
| ---------
| ---------
| ---------
|
| L
| w/Y; 255B/+;
Pk61Ca/+ | M
| 21
| 42.81±13.89
| 47
| -10.63
| 0
| 0.224
|
| -
| w/yw; 255B/+;
Pk61Ca/+ | F
| 127
| 49.82±16.97
| 56
| ---------
| ---------
| ---------
|
| L
| w/yw; 255B/+;
Pk61Ca/+ | F
| 121
| 50.24±11.55
| 52
| 0.84
| -7.14
| 8.57E-04
|
| 18-1
| -
| w/Y; 255B/+;
Pk61Cb/+ | M
| 126
| 57.29±8.23
| 59
| ---------
| ---------
| ---------
|
| L
| w/Y; 255B/+;
Pk61Cb/+ | M
| 24
| 48.08±10.99
| 51
| -16.06
| -13.56
| 4.11E-12
|
| -
| w; 255B/+;
Pk61Cb/+ | F
| 124
| 54.51±9.89
| 56.5
| ---------
| ---------
| ---------
|
| L
| w; 255B/+;
Pk61Cb/+ | F
| 121
| 45.28±13.38
| 51
| -16.93
| -9.73
| 1.79E-14
|
| 19-1
| -
| w/Y; 255B/+;
DIAP1/+ | M
| 120
| 56.23±8.68
| 59
| ---------
| ---------
| ---------
|
| L
| w/Y; 255B/+;
DIAP1/+ | M
| 93
| 47.68±17.07
| 53
| -15.22
| -10.17
| 0.002
|
| -
| w/w*; 255B/+;
DIAP1/+ | F
| 120
| 54.64±11.2
| 57.5
| ---------
| ---------
| ---------
|
| L
| w/w*; 255B/+;
DIAP1/+ | F
| 117
| 46.74±14.76
| 51
| -14.45
| -11.3
| 1.97E-07
|
| 7-1
| -
| w/Y; 255B/+;
+ | M
| 124
| 52.97±7.68
| 56
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
+ | M
| 124
| 49.73±4.6
| 50
| -6.11
| -10.71
| 6.19E-13
|
| L
| w/Y; 255B/+;
+ | M
| 22
| 44.5±16.86
| 52
| -15.99
| -7.14
| 2.94E-05
|
| -
| w/+; 255B/+;
+ | F
| 118
| 58.39±5.25
| 59
| ---------
| ---------
| ---------
|
| A
| w/+; 255B/+;
+ | F
| 122
| 52.66±4.09
| 52
| -9.81
| -11.86
| 0
|
| L
| w/+; 255B/+;
+ | F
| 122
| 50.45±9.78
| 53.5
| -13.6
| -9.32
| 1.18E-13
|
| Ex 2 Life
span assay using GS255B driver at 25C |
| 7-1
| -
| w/Y; 255B/+;
+ | M
| 94
| 73.17±15.64
| 78
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
+ | M
| 93
| 69.97±12.33
| 72
| -4.38
| -7.69
| 5.22E-04
|
| -
| w/+; 255B/+;
+ | F
| 92
| 87.2±18.44
| 92
| ---------
| ---------
| ---------
|
| A
| w/+; 255B/+;
+ | F
| 91
| 91.93±7.76
| 94
| 5.43
| 2.17
| 0.940
|
| 20-1
| -
| w/Y; 255B/+;
nmo/+ | M
| 95
| 66.74±16.11
| 68
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
nmo/+ | M
| 90
| 64.66±14.3
| 66
| -3.12
| -2.94
| 0.102
|
| -
| w/yw; 255B/+;
nmo/+ | F
| 97
| 67.59±28.66
| 74
| ---------
| ---------
| ---------
|
| A
| w/yw; 255B/+;
nmo/+ | F
| 95
| 68.79±30.17
| 80
| 1.78
| 8.11
| 0.878
|
| 15-1
| -
| w/Y; 255B/wga;
+ | M
| 96
| 72.88±10.31
| 74
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/wga;
+ | M
| 92
| 53.14±18.01
| 56
| -27.08
| -24.32
| 0
|
| -
| w/w*; 255B/wga;
+ | F
| 97
| 78.89±19.38
| 84
| ---------
| ---------
| ---------
|
| A
| w/w*; 255B/wga;
+ | F
| 97
| 53.72±22.13
| 52
| -31.9
| -38.1
| 0
|
| 17-1
| -
| w/Y; 255B/+;
Pk61Ca/+ | M
| 91
| 64.11±13.4
| 64
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
Pk61Ca/+ | M
| 94
| 62.85±13.08
| 66
| -1.96
| 3.13
| 0.555
|
| -
| w/yw; 255B/+;
Pk61Ca/+ | F
| 98
| 70.73±26.23
| 78
| ---------
| ---------
| ---------
|
| A
| w/yw; 255B/+;
Pk61Ca/+ | F
| 94
| 79.81±23.77
| 90
| 12.83
| 15.38
| 0.149
|
| 21-1
| -
| w/Y; 255B/+;
Egfr/+ | M
| 89
| 62.38±11.19
| 66
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
Egfr/+ | M
| 97
| 62.06±9.18
| 64
| -0.51
| -3.03
| 0.166
|
| -
| w/y w*;
255B/+; Egfr/+ | F
| 95
| 65.71±21.16
| 68
| ---------
| ---------
| ---------
|
| A
| w/y w*;
255B/+; Egfr/+ | F
| 100
| 63.52±17.9
| 65
| -3.33
| -4.41
| 0.076
|
| 19-1
| -
| w/Y; 255B/+;
DIAP1/+ | M
| 102
| 76.57±13.04
| 78
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
DIAP1/+ | M
| 94
| 73.4±9.53
| 74
| -4.13
| -5.13
| 0.002
|
| -
| w/w*; 255B/+;
DIAP1/+ | F
| 98
| 78.9±18.26
| 84
| ---------
| ---------
| ---------
|
| A
| w/w*; 255B/+;
DIAP1/+ | F
| 95
| 81.39±19.17
| 88
| 3.16
| 4.76
| 0.011
|
| 22-1
| -
| w/Y; 255B/+;
pnt/+ | M
| 96
| 62.6±9.74
| 64
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
pnt/+ | M
| 94
| 59.15±10.7
| 60
| -5.52
| -6.25
| 0.077
|
| -
| w/yw; 255B/+;
pnt/+ | F
| 92
| 74.32±27.19
| 85
| ---------
| ---------
| ---------
|
| A
| w/yw; 255B/+;
pnt/+ | F
| 95
| 79.77±20.03
| 88
| 7.34
| 3.53
| 0.402
|
| Exp3 Life
span assay using GS255B driver at 25C |
| 7-1
| -
| w/Y; 255B/+;
+ | M
| 100
| 81.01±15.38
| 86
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
+ | M
| 92
| 80.46±10.42
| 82
| -0.68
| -4.65
| 0.039
|
| -
| w/+; 255B/+;
+ | F
| 85
| 92.49±11.86
| 94
| ---------
| ---------
| ---------
|
| A
| w/+; 255B/+;
+ | F
| 99
| 92.05±13.07
| 94
| -0.48
| 0
| 0.571
|
| 13-1
| -
| w/Y; 255B/Ras
WT;+ | M
| 95
| 75.87±12.26
| 78
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/Ras
WT;+ | M
| 98
| 67.92±14.13
| 70
| -10.48
| -10.26
| 1.62E-06
|
| -
| w/w*;
255B/Ras WT;+ | F
| 96
| 83.43±12.26
| 86
| ---------
| ---------
| ---------
|
| A
| w/w*;
255B/Ras WT;+ | F
| 98
| 79.59±10.58
| 82
| -4.6
| -4.65
| 0.001
|
| 23-1
| -
| w/Y;
255B/Mmp2; + | M
| 96
| 69.77±13.03
| 70
| ---------
| ---------
| ---------
|
| A
| w/Y;
255B/Mmp2; + | M
| 96
| 68.81±10.06
| 70
| -1.37
| 0
| 0.117
|
| -
| w/yw;
255B/Mmp2; + | F
| 98
| 85.94±18.45
| 91
| ---------
| ---------
| ---------
|
| A
| w/yw;
255B/Mmp2; + | F
| 101
| 84.85±18.67
| 90
| -1.27
| -1.1
| 0.109
|
| 24-1
| -
| w/Y; 255B/+;
Stat/+ | M
| 96
| 64.31±10.06
| 65
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
Stat/+ | M
| 99
| 65.08±13.33
| 68
| 1.19
| 4.62
| 0.325
|
| -
| w/yw; 255B/+;
Stat/+ | F
| 99
| 70.48±21.48
| 78
| ---------
| ---------
| ---------
|
| A
| w/yw; 255B/+;
Stat/+ | F
| 96
| 62.29±24.99
| 74
| -11.62
| -5.13
| 0.076
|
| 25-1
| -
| w/Y; 255B/+;
puc/+ | M
| 97
| 70.78±14.98
| 70
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
puc/+ | M
| 96
| 68.96±13.62
| 68
| -2.58
| -2.86
| 0.269
|
| -
| w/w*; 255B/+;
puc/+ | F
| 84
| 94.07±15.00
| 98
| ---------
| ---------
| ---------
|
| A
| w/w*; 255B/+;
puc/+ | F
| 97
| 98.1±7.86
| 100
| 4.29
| 2.04
| 0.135
|
| Exp4 Life
span assay using GS255B driver at 25C |
| 7-1
| -
| w/Y; 255B/+;
+ | M
| 92
| 73.76±18.31
| 78
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
+ | M
| 86
| 71.84±10.87
| 74
| -2.61
| -5.13
| 1.43E-04
|
| -
| w/+; 255B/+;
+ | F
| 86
| 86.28±15.46
| 90
| ---------
| ---------
| ---------
|
| A
| w/+; 255B/+;
+ | F
| 101
| 86.18±10.02
| 88
| -0.12
| -2.22
| 0.035
|
| 26-1
| -
| w/Y; 255B/+;
Sk2/+ | M
| 90
| 67.22±16.97
| 72
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
Sk2/+ | M
| 95
| 69.85±12.26
| 72
| 3.91
| 0
| 0.953
|
| -
| w/yw; 255B/+;
Sk2/+ | F
| 101
| 73.29±24.6
| 84
| ---------
| ---------
| ---------
|
| A
| w/yw; 255B/+;
Sk2/+ | F
| 106
| 78.08±19.61
| 86
| 6.53
| 2.38
| 0.84
|
| 27-1
| -
| w/Y; 255B/+;
ban/+ | M
| 98
| 66.59±21.91
| 70
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
ban/+ | M
| 95
| 61.56±18.15
| 62
| -7.56
| -11.43
| 0.003
|
| -
| w/yw; 255B/+;
ban/+ | F
| 94
| 76.36±28.78
| 88
| ---------
| ---------
| ---------
|
| A
| w/yw; 255B/+;
ban/+ | F
| 96
| 81.56±18.03
| 88
| 6.81
| 0
| 0.023
|
| 28-1
| -
| w/Y;
255B/+;CG14544/+ | M
| 91
| 75.03±12.8
| 76
| ---------
| ---------
| ---------
|
| A
| w/Y;
255B/+;CG14544/+ | M
| 97
| 77.01±9.82
| 78
| 2.64
| 2.63
| 0.844
|
| -
| w/w;
255B/+;CG14544/+ | F
| 101
| 70.99±26.75
| 82
| ---------
| ---------
| ---------
|
| A
| w/w;
255B/+;CG14544/+ | F
| 96
| 69.33±20.57
| 79
| -2.33
| -3.66
| 2.28E-05
|
| Exp5 Life
span assay using GS255B driver, and MHC GS driver at 25C |
| 7-1
| -
| w/Y; 255B/+;
+ | M
| 98
| 75.06±11.65
| 79
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
+ | M
| 97
| 73.96±12.27
| 78
| -1.47
| -1.27
| 0.161
|
| -
| w/+; 255B/+;
+ | F
| 100
| 87.88±7.74
| 88
| ---------
| ---------
| ---------
|
| A
| w/+; 255B/+;
+ | F
| 101
| 85.33±12.78
| 88
| -2.91
| 0
| 0.014
|
| 8-1
| -
| w/Y; 255B/+;
+ | M
| 99
| 66.55±11.82
| 68
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
+ | M
| 97
| 69.84±10.57
| 72
| 4.94
| 5.88
| 0.14
|
| -
| w/w; 255B/+;
+ | F
| 100
| 79.6±14.08
| 84
| ---------
| ---------
| ---------
|
| A
| w/w; 255B/+;
+ | F
| 97
| 81.69±3.82
| 82
| 2.63
| -2.38
| 0.005
|
| 17-1
| -
| w/Y; 255B/+;
Pk61Ca/+ | M
| 99
| 64.87±12.41
| 64
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
Pk61Ca/+ | M
| 98
| 62.16±12.33
| 64
| -4.17
| 0
| 0.113
|
| -
| w/yw; 255B/+;
Pk61Ca/+ | F
| 98
| 81.96±13.91
| 86
| ---------
| ---------
| ---------
|
| A
| w/yw; 255B/+;
Pk61Ca/+ | F
| 99
| 80.46±11.15
| 82
| -1.82
| -4.65
| 0.001
|
| 18-1
| -
| w/Y; 255B/+;
Pk61Cb/+ | M
| 100
| 71.2±9.32
| 74
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
Pk61Cb/+ | M
| 101
| 73.33±8.19
| 74
| 2.99
| 0
| 0.135
|
| -
| w; 255B/+;
Pk61Cb/+ | F
| 98
| 80.27±12.63
| 82
| ---------
| ---------
| ---------
|
| A
| w; 255B/+;
Pk61Cb/+ | F
| 100
| 82.02±4.89
| 82
| 2.19
| 0
| 0.399
|
| 19-1
| -
| w/Y; 255B/+;
DIAP1/+ | M
| 101
| 72.97±11.13
| 76
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;
DIAP1/+ | M
| 101
| 71.15±11.37
| 74
| -2.5
| -2.63
| 0.425
|
| -
| w/w*; 255B/+;
DIAP1/+ | F
| 98
| 83.02±7.52
| 84
| ---------
| ---------
| ---------
|
| A
| w/w*; 255B/+;
DIAP1/+ | F
| 106
| 79.47±12.78
| 82
| -4.27
| -2.38
| 0.011
|
| 7-6
| -
| yw/Y;+/CyO;
MHC/+ | M
| 96
| 71.31±13.18
| 76
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/CyO;
MHC/+ | M
| 98
| 58.92±12.84
| 60
| -17.38
| -21.05
| 0
|
| -
| yw/+;+/CyO;
MHC/+ | F
| 96
| 78.04±15.76
| 84
| ---------
| ---------
| ---------
|
| A
| yw/+;+/CyO;
MHC/+ | F
| 114
| 52.84±13.75
| 58
| -32.29
| -30.95
| 0
|
| 8-6
| -
| yw/Y;+/CyO;
MHC/+ | M
| 99
| 56.42±16.69
| 60
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/CyO;
MHC/+ | M
| 92
| 43.26±15
| 48
| -23.33
| -20
| 1.61E-12
|
| -
| yw/w;+/CyO;
MHC/+ | F
| 93
| 60.73±18.82
| 68
| ---------
| ---------
| ---------
|
| A
| yw/w;+/CyO;
MHC/+ | F
| 99
| 45.21±13.63
| 50
| -25.55
| -26.47
| 2.22E-16
|
| 17-6
| -
| yw/Y;+/CyO;
MHC/Pk61Ca | M
| 100
| 54.32±14.66
| 58
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/CyO;
MHC/Pk61Ca | M
| 101
| 52.46±12.42
| 56
| -3.43
| -3.45
| 0.004
|
| -
| yw;+/CyO;
MHC/Pk61Ca | F
| 100
| 61.3±19.84
| 65
| ---------
| ---------
| ---------
|
| A
| yw;+/CyO;
MHC/Pk61Ca | F
| 99
| 39.37±14.22
| 36
| -35.77
| -44.62
| 0
|
| 18-6
| -
| yw/Y;+/CyO;
MHC/Pk61Cb | M
| 96
| 56.44±18.89
| 62
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/CyO;
MHC/Pk61Cb | M
| 108
| 53.56±10.21
| 56
| -5.11
| -9.68
| 1.75E-07
|
| -
| yw/w;+/CyO;
MHC/Pk61Cb | F
| 96
| 55.25±21.21
| 56
| ---------
| ---------
| ---------
|
| A
| yw/w;+/CyO;
MHC/Pk61Cb | F
| 90
| 39.42±12.45
| 38
| -28.65
| -32.14
| 1.40E-11
|
| 19-6
| -
| yw/Y;+/CyO;
MHC/DIAP1 | M
| 98
| 66.71±12.27
| 68
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/CyO;
MHC/DIAP1 | M
| 98
| 54.92±10.4
| 56
| -17.68
| -17.65
| 6.66E-15
|
| -
| yw/w*;+/CyO;MHC/DIAP1 | F
| 108
| 64.89±18.19
| 71
| ---------
| ---------
| ---------
|
| A
| yw/w*;+/CyO;MHC/DIAP1 | F
| 104
| 57.04±14.85
| 58
| -12.1
| -18.31
| 6.72E-08
|
| 7-6
| -
| yw/Y;+/Sp;
MHC/+ | M
| 99
| 68.67±13.38
| 74
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/Sp;
MHC/+ | M
| 98
| 61.76±12.62
| 64
| -10.07
| -13.51
| 5.34E-08
|
| -
| yw/+;+/Sp;
MHC/+ | F
| 94
| 73.66±13.99
| 78
| ---------
| ---------
| ---------
|
| A
| yw/+;+/Sp;
MHC/+ | F
| 102
| 59.98±8.45
| 62
| -18.57
| -20.51
| 0
|
| 8-6
| -
| yw/Y;+/Sp;
MHC/+ | M
| 96
| 61.6±14.81
| 66
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/Sp;
MHC/+ | M
| 94
| 54.55±15.5
| 56
| -11.45
| -15.15
| 2.47E-06
|
| -
| yw/w;+/Sp;
MHC/+ | F
| 93
| 58.41±17.34
| 62
| ---------
| ---------
| ---------
|
| A
| yw/w;+/Sp;
MHC/+ | F
| 102
| 47.59±12.97
| 52
| -18.53
| -16.13
| 2.70E-14
|
| 17-6
| -
| yw/Y;+/Sp;
MHC/Pk61Ca | M
| 95
| 49.31±12.75
| 54
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/Sp;
MHC/Pk61Ca | M
| 100
| 45.14±11.03
| 48
| -8.45
| -11.11
| 1.95E-04
|
| -
| yw;+/Sp;
MHC/Pk61Ca | F
| 99
| 42.99±20.17
| 40
| ---------
| ---------
| ---------
|
| A
| yw;+/Sp;
MHC/Pk61Ca | F
| 100
| 35.78±16.13
| 30
| -16.77
| -25
| 0.003
|
| 18-6
| -
| yw/Y;+/Sp;
MHC/Pk61Cb | M
| 100
| 56.36±12.56
| 60
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/Sp;
MHC/Pk61Cb | M
| 98
| 51.06±9.77
| 52
| -9.4
| -13.33
| 2.13E-06
|
| -
| yw/w;+/Sp;
MHC/Pk61Cb | F
| 97
| 56.6±16.25
| 60
| ---------
| ---------
| ---------
|
| A
| yw/w;+/Sp;
MHC/Pk61Cb | F
| 94
| 41.79±11.81
| 42
| -26.17
| -30
| 1.67E-15
|
| 19-6
| -
| yw/Y;+/Sp;
MHC/DIAP1 | M
| 99
| 61.21±10.04
| 62
| ---------
| ---------
| ---------
|
| A
| yw/Y;+/Sp;
MHC/DIAP1 | M
| 97
| 55.51±9.17
| 58
| -9.32
| -6.45
| 5.02E-08
|
| -
| yw/w*;+/Sp;
MHC/DIAP1 | F
| 103
| 71.13±17.02
| 78
| ---------
| ---------
| ---------
|
| A
| yw/w*;+/Sp;
MHC/DIAP1 | F
| 101
| 62.3±13.23
| 66
| -12.41
| -15.38
| 6.66E-16
|
Table 3. Life span data for baculovirus p35 experiments, with means, standard
deviations, medians, percent change in mean and median, and log rank p
value.
| CrossMxF | RU486 | Genotype | Sex | N | Meana | Median | %Change in Mean | %Change in Median | Log Rank p Value |
| Exp1 Life span assay of
three UAS-p35 lines and UAS-p53.259H with GS255B driver at 25C |
| 7-1
| -
| w/Y; 255B/+; + | M
| 120
| 84.6±14.25
| 90
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+; + | M
| 119
| 83.08±10.94
| 86
| -1.8
| -4.44
| 0.014
|
| L
| w/Y; 255B/+; + | M
| 123
| 78.44±22.48
| 86
| -7.28
| -4.44
| 0.244
|
| -
| w/+; 255B/+; + | F
| 116
| 92.02±9.64
| 94
| ---------
| ---------
| ---------
|
| A
| w/+; 255B/+; + | F
| 121
| 94.69±8.61
| 94
| 2.9
| 0
| 0.009
|
| L
| w/+; 255B/+; + | F
| 124
| 91.97±15.74
| 94
| -0.05
| 0
| 0.047
|
| 1-9
| -
| p35,w*/Y; 255B/+; + | M
| 120
| 68.93±12.49
| 70
| ---------
| ---------
| ---------
|
| A
| p35,w*/Y; 255B/+; + | M
| 123
| 68.62±11.76
| 70
| -0.45
| 0
| 0.681
|
| L
| p35,w*/Y; 255B/+; + | M
| 98
| 33.1±22.91
| 26
| -51.98
| -62.86
| 0
|
| -
| p35,w*/+; 255B/+; + | F
| 122
| 83.28±15.13
| 86
| ---------
| ---------
| ---------
|
| A
| p35,w*/+; 255B/+; + | F
| 130
| 77.15±20.92
| 82
| -7.36
| -4.65
| 0.11
|
| L
| p35,w*/+; 255B/+; + | F
| 125
| 57.52±35.8
| 70
| -30.93
| -18.6
| 0.001
|
| 10-1
| -
| w/Y; 255B/p35; + | M
| 117
| 54.48±13
| 54
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/p35; + | M
| 121
| 57.26±8.45
| 58
| 5.1
| 7.41
| 0.583
|
| L
| w/Y; 255B/p35; + | M
| 110
| 34.25±19.5
| 34
| -37.13
| -37.04
| 5.60E-10
|
| -
| w/w*; 255B/p35; + | F
| 120
| 64.05±14.63
| 66
| ---------
| ---------
| ---------
|
| A
| w/w*; 255B/p35; + | F
| 126
| 60.79±16.68
| 66
| -5.09
| 0
| 0.188
|
| L
| w/w*; 255B/p35; + | F
| 123
| 49.37±32.2
| 54
| -22.92
| -18.18
| 0.436
|
| 11-1
| -
| w/Y; 255B/+; p35/+ | M
| 133
| 86.03±12.51
| 90
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+; p35/+ | M
| 122
| 81.31±13.87
| 84
| -5.49
| -6.67
| 8.92E-06
|
| L
| w/Y; 255B/+; p35/+ | M
| 56
| 46.18±24.21
| 44
| -46.32
| -51.11
| 0
|
| -
| w/w*; 255B/+; p35/+ | F
| 126
| 87.54±10.04
| 90
| ---------
| ---------
| ---------
|
| A
| w/w*; 255B/+; p35/+ | F
| 127
| 82.19±9.91
| 82
| -6.11
| -8.89
| 8.60E-05
|
| L
| w/w*; 255B/+; p35/+ | F
| 126
| 64.63±29.62
| 75
| -26.17
| -16.67
| 4.67E-07
|
| 29-1
| -
| w/Y; 255B/+;p53.259H/+ | M
| 118
| 71.54±13.86
| 72
| ---------
| ---------
| ---------
|
| A
| w/Y; 255B/+;p53.259H/+ | M
| 125
| 68.90±10.41
| 70
| -3.70
| -2.78
| 0.002
|
| L
| w/Y; 255B/+;p53.259H/+ | M
| 119
| 67.73±16.92
| 70
| -5.33
| -2.78
| 0.069
|
| -
| w; 255B/+; p53.259H/+ | F
| 119
| 75.40±8.50
| 76
| ---------
| ---------
| ---------
|
| A
| w; 255B/+; p53.259H/+ | F
| 119
| 80.66±10.98
| 82
| 6.98
| 7.89
| 4.05E-08
|
| L
| w; 255B/+; p53.259H/+ | F
| 125
| 70.24±22.02
| 76
| -6.84
| 0
| 0.202
|
| Exp2 Life span assay of
three UAS-p35 lines with head FB driver, whole body FB driver and GS255A
driver at 25C |
| 3-7
| -
| +/Y; S32/+; + | M
| 75
| 59.23±14.11
| 64
| ---------
| ---------
| ---------
|
| A
| +/Y; S32/+; + | M
| 63
| 55.21±15.74
| 60
| -6.79
| -6.25
| 0.013
|
| -
| w/+; S32/+; + | F
| 111
| 59.91±18.96
| 60
| ---------
| ---------
| ---------
|
| A
| w/+; S32/+; + | F
| 115
| 63.77±17.71
| 66
| 6.45
| 10
| 0.263
|
| 3-9
| -
| p35,w*/Y; S32/+; + | M
| 122
| 62.69±10.62
| 64
| ---------
| ---------
| ---------
|
| A
| p35,w*/Y; S32/+; + | M
| 105
| 58.3±13.45
| 60
| -6.99
| -6.25
| 0.022
|
| -
| p35,w*/w; S32/+; + | F
| 112
| 59.95±25
| 72
| ---------
| ---------
| ---------
|
| A
| p35,w*/w; S32/+; + | F
| 108
| 59±24.86
| 68
| -1.58
| -5.56
| 0.974
|
| 3-10
| -
| w*/Y; S32/p35; + | M
| 123
| 45.19±7.61
| 46
| ---------
| ---------
| ---------
|
| A
| w*/Y; S32/p35; + | M
| 120
| 41.52±6.84
| 42
| -8.12
| -8.7
| 1.96E-04
|
| -
| w*/w; S32/p35; + | F
| 121
| 61.62±8.71
| 62
| ---------
| ---------
| ---------
|
| A
| w*/w; S32/p35; + | F
| 105
| 63.28±10.6
| 66
| 2.69
| 6.45
| 0.036
|
| 3-11
| -
| w*/Y; S32/+; p35/+ | M
| 125
| 62.67±12.41
| 64
| ---------
| ---------
| ---------
|
| A
| w*/Y; S32/+; p35/+ | M
| 125
| 60.78±14.07
| 62
| -3.03
| -3.13
| 0.174
|
| -
| w*/w; S32/+; p35/+ | F
| 109
| 68.44±17.09
| 74
| ---------
| ---------
| ---------
|
| A
| w*/w; S32/+; p35/+ | F
| 113
| 70.52±18.12
| 76
| 3.04
| 2.7
| 0.043
|
| 4-7
| -
| +/Y; S106 S32/+; + | M
| 116
| 54.12±9.89
| 56
| ---------
| ---------
| ---------
|
| A
| +/Y; S106 S32/+; + | M
| 118
| 52.68±9.77
| 52
| -2.67
| -7.14
| 0.208
|
| -
| w/+; S106 S32/+; + | F
| 110
| 58.82±14.95
| 62
| ---------
| ---------
| ---------
|
| A
| w/+; S106 S32/+; + | F
| 120
| 58.07±16.45
| 63
| -1.28
| 1.61
| 0.569
|
| 4-9
| -
| p35,w*/Y; S106 S32/+; + | M
| 121
| 47.21±8.48
| 46
| ---------
| ---------
| ---------
|
| A
| p35,w*/Y; S106 S32/+; + | M
| 110
| 47.67±10.28
| 48
| 0.99
| 4.35
| 0.263
|
| -
| p35,w*/w; S106 S32/+; + | F
| 119
| 55.18±22.95
| 66
| ---------
| ---------
| ---------
|
| A
| p35,w*/w; S106 S32/+; + | F
| 126
| 47.79±24.9
| 62
| -13.38
| -6.06
| 0.01
|
| 4-10
| -
| w*/Y; S106 S32/p35; + | M
| 125
| 33.39±4.44
| 34
| ---------
| ---------
| ---------
|
| A
| w*/Y; S106 S32/p35; + | M
| 125
| 32.3±6.16
| 32
| -3.26
| -5.88
| 0.475
|
| -
| w*/w; S106 S32/p35; + | F
| 121
| 49.55±8.14
| 50
| ---------
| ---------
| ---------
|
| A
| w*/w; S106 S32/p35; + | F
| 121
| 50.84±8.85
| 50
| 2.6
| 0
| 0.107
|
| 4-11
| -
| w*/Y; S106 S32/+; p35/+ | M
| 125
| 47.15±6.81
| 48
| ---------
| ---------
| ---------
|
| A
| w*/Y; S106 S32/+; p35/+ | M
| 117
| 48.6±8.42
| 48
| 3.07
| 0
| 0.072
|
| -
| w*/w; S106 S32/+; p35/+ | F
| 125
| 56.81±13.02
| 60
| ---------
| ---------
| ---------
|
| A
| w*/w; S106 S32/+; p35/+ | F
| 116
| 60.69±11.7
| 64
| 6.83
| 6.67
| 0.004
|
| 2-7
| -
| +/Y; 255A/+; + | M
| 114
| 66.04±8.95
| 67
| ---------
| ---------
| ---------
|
| A
| +/Y; 255A/+; + | M
| 117
| 58.97±15.36
| 62
| -10.69
| -7.46
| 1.48E-05
|
| -
| w/+; 255A/+; + | F
| 114
| 72.65±13.95
| 78
| ---------
| ---------
| ---------
|
| A
| w/+; 255A/+; + | F
| 116
| 75.02±13.19
| 78
| 3.26
| 0
| 0.064
|
| 2-9
| -
| p35,w*/Y; 255A/+; + | M
| 111
| 65.98±14.65
| 66
| ---------
| ---------
| ---------
|
| A
| p35,w*/Y; 255A/+; + | M
| 115
| 59.82±13.31
| 60
| -9.34
| -9.09
| 3.78E-05
|
| -
| p35,w*/w; 255A/+; + | F
| 113
| 58.95±20.26
| 64
| ---------
| ---------
| ---------
|
| A
| p35,w*/w; 255A/+; + | F
| 117
| 69.21±17.4
| 72
| 17.4
| 12.5
| 1.32E-06
|
| 2-10
| -
| w*/Y;255A/p35; + | M
| 113
| 48.98±9.74
| 48
| ---------
| ---------
| ---------
|
| A
| w*/Y;255A/p35; + | M
| 125
| 47.66±7.19
| 48
| -2.69
| 0
| 0.03
|
| -
| w*/w; 255A/p35; + | F
| 115
| 60.57±16.71
| 66
| ---------
| ---------
| ---------
|
| A
| w*/w; 255A/p35; + | F
| 118
| 62±17.79
| 70
| 2.35
| 6.06
| 0.052
|
| 2-11
| -
| w*/Y; 255A/+; p35/+ | M
| 115
| 63.66±11.4
| 64
| ---------
| ---------
| ---------
|
| A
| w*/Y; 255A/+; p35/+ | M
| 114
| 64.92±9.41
| 64
| 1.98
| 0
| 0.776
|
| -
| w*/w; 255A/+; p35/+ | F
| 120
| 67.05±11.58
| 70
| ---------
| ---------
| ---------
|
| A
| w*/w; 255A/+; p35/+ | F
| 120
| 68.75±9.08
| 70
| 2.54
| 0
| 0.41
|
| Exp3 Life span assay of
two UAS-p35 lines with whole body FB driver at 29C |
| 7-4
| -
| w/Y; S106 S32/+; + | M
| 124
| 49.15±12.5
| 54
| ---------
| ---------
| ---------
|
| L
| w/Y; S106 S32/+; + | M
| 121
| 49.11±10.75
| 52
| -0.08
| -3.7
| 0.655
|
| -
| w/+; S106 S32/+; + | F
| 121
| 51.95±10.82
| 54
| ---------
| ---------
| ---------
|
| L
| w/+; S106 S32/+; + | F
| 118
| 55.29±10.06
| 60
| 6.42
| 11.11
| 0.029
|
| 8-4
| -
| w/Y; S106 S32/+; + | M
| 121
| 47.16±10.27
| 48
| ---------
| ---------
| ---------
|
| L
| w/Y; S106 S32/+; + | M
| 118
| 42.85±12.89
| 44
| -9.14
| -8.33
| 0.002
|
| -
| w/w; S106 S32/+; + | F
| 124
| 50.48±11.91
| 56
| ---------
| ---------
| ---------
|
| L
| w/w; S106 S32/+; + | F
| 125
| 51.63±8.47
| 54
| 2.27
| -3.57
| 0.196
|
| 10-4
| -
| w/Y; S106 S32/p35; + | M
| 121
| 50.43±6.73
| 52
| ---------
| ---------
| ---------
|
| L
| w/Y; S106 S32/p35; + | M
| 121
| 46.5±8.28
| 48
| -7.8
| -7.69
| 4.23E-05
|
| -
| w*/w; S106 S32/p35; + | F
| 120
| 50.4±12.84
| 56
| ---------
| ---------
| ---------
|
| L
| w*/w; S106 S32/p35; + | F
| 129
| 48.57±8.9
| 50
| -3.62
| -10.71
| 1.45E-05
|
| 11-4
| -
| w/Y; S106 S32/+; p35/+ | M
| 126
| 44.03±6.5
| 46
| ---------
| ---------
| ---------
|
| L
| w/Y; S106 S32/+; p35/+ | M
| 122
| 41.92±10.42
| 46
| -4.8
| 0
| 0.208
|
| -
| w*/w; S106 S32/+; p35/+ | F
| 122
| 58.03±6.59
| 60
| ---------
| ---------
| ---------
|
| L
| w*/w; S106 S32/+; p35/+ | F
| 124
| 54.81±8.05
| 56
| -5.56
| -6.67
| 8.84E-07
|
| Exp4 Life span assay of
two UAS-p35 lines with Elav driver at 29C |
| 7-5
| -
| yw/Y; +/+; Elav/+ | M
| 131
| 53.92±7.15
| 54
| ---------
| ---------
| ---------
|
| A
| yw/Y; +/+; Elav/+ | M
| 129
| 52.33±8.14
| 53
| -2.95
| -1.85
| 0.083
|
| L
| yw/Y; +/+; Elav/+ | M
| 59
| 35.85±10.58
| 38
| -33.51
| -29.63
| 0
|
| -
| yw/+; +/+; Elav/+ | F
| 127
| 58.15±7.22
| 60
| ---------
| ---------
| ---------
|
| A
| yw/+; +/+; Elav/+ | F
| 129
| 57.11±5.19
| 58
| -1.79
| -3.33
| 0.013
|
| L
| yw/+; +/+; Elav/+ | F
| 120
| 43.46±7.94
| 44
| -25.26
| -26.67
| 0
|
| 8-5
| -
| yw/Y; +/+; Elav/+ | M
| 126
| 44.08±8.36
| 44.5
| ---------
| ---------
| ---------
|
| A
| yw/Y; +/+; Elav/+ | M
| 120
| 43.32±8.76
| 45
| -1.73
| 1.12
| 0.186
|
| L
| yw/Y; +/+; Elav/+ | M
| 102
| 26.24±8.51
| 26
| -40.48
| -41.57
| 0
|
| -
| yw/w; +/+; Elav/+ | F
| 124
| 46.88±9.92
| 50
| ---------
| ---------
| ---------
|
| A
| yw/w; +/+; Elav/+ | F
| 124
| 48.13±7.5
| 49.5
| 2.67
| -1
| 0.406
|
| L
| yw/w; +/+; Elav/+ | F
| 114
| 34.82±10.34
| 36
| -25.73
| -28
| 0
|
| 10-5
| -
| yw/Y; p35/+; Elav/+ | M
| 125
| 42.34±6.38
| 44
| ---------
| ---------
| ---------
|
| A
| yw/Y; p35/+; Elav/+ | M
| 122
| 43.34±10.34
| 46
| 2.38
| 4.55
| 0.007
|
| L
| yw/Y; p35/+; Elav/+ | M
| 9
| 20.89±10.3
| 26
| -50.66
| -40.91
| 0
|
| -
| yw/w*; p35/+; Elav/+ | F
| 121
| 49±10.63
| 52
| ---------
| ---------
| ---------
|
| A
| yw/w*; p35/+; Elav/+ | F
| 126
| 50.16±6.4
| 51
| 2.36
| -1.92
| 0.014
|
| L
| yw/w*; p35/+; Elav/+ | F
| 9
| 28.22±10.27
| 32
| -42.4
| -38.46
| 1.60E-14
|
| 11-5
| -
| yw/Y; +; Elav/p35 | M
| 120
| 51.24±10.46
| 54
| ---------
| ---------
| ---------
|
| A
| yw/Y; +; Elav/p35 | M
| 121
| 48.62±10.1
| 52
| -5.12
| -3.7
| 1.22E-06
|
| L
| yw/Y; +; Elav/p35 | M
| 1
| 10±NA
| 10
| -80.48
| -81.48
| 5.60E-10
|
| -
| yw/w*; +; Elav/p35 | F
| 118
| 56.77±3.89
| 58
| ---------
| ---------
| ---------
|
| A
| yw/w*; +; Elav/p35 | F
| 131
| 52.67±5.08
| 54
| -7.22
| -6.9
| 3.51E-13
|
| L
| yw/w*; +; Elav/p35 | F
| 0
| NA
| NA
| ---------
| ---------
| ---------
|
| Exp5 Life span assay of
two UAS-p35 lines with GS255B driver at 25C |
| 8-1
| -
| w/Y; 255B/+; + | M
| 121
| 62.33±18.12
| 68
| ---------
| ---------
| ---------
|
| L
| w/Y; 255B/+; + | M
| 119
| 62.57±16.22
| 68
| 0.39
| 0
| 0.478
|
| L1-10
| w/Y; 255B/+; + | M
| 120
| 66.02±19.38
| 72
| 5.91
| 5.88
| 7.82E-04
|
| -
| w/w; 255B/+; + | F
| 123
| 75.95±9.37
| 78
| ---------
| ---------
| ---------
|
| L
| w/w; 255B/+; + | F
| 124
| 69.02±12.88
| 74
| -9.13
| -5.13
| 7.69E-07
|
| L1-10
| w/w; 255B/+; + | F
| 124
| 78.18±9.17
| 80
| 2.93
| 2.56
| 7.84E-04
|
| 10-1
| -
| w/Y; 255B/p35; + | M
| 111
| 56.32±25.51
| 66
| ---------
| ---------
| ---------
|
| L
| w/Y; 255B/p35; + | M
| 4
| 16±21.6
| 7
| -71.59
| -89.39
| 6.47E-05
|
| L1-10
| w/Y; 255B/p35; + | M
| 117
| 58.56±17.95
| 62
| 3.98
| -6.06
| 0.528
|
| -
| w/w*; 255B/p35; + | F
| 119
| 68.47±13.26
| 72
| ---------
| ---------
| ---------
|
| L
| w/w*; 255B/p35; + | F
| 30
| 27.47±16.58
| 24
| -59.89
| -66.67
| 0
|
| L1-10
| w/w*; 255B/p35; + | F
| 124
| 64.5±16.45
| 70
| -5.8
| -2.78
| 0.757
|
| 11-1
| -
| w/Y; 255B/+; p35/+ | M
| 117
| 66.15±9.97
| 68
| ---------
| ---------
| ---------
|
| L
| w/Y; 255B/+; p35/+ | M
| 1
| 14±NA
| 14
| -78.84
| -79.41
| 3.38E-14
|
| L1-10
| w/Y; 255B/+; p35/+ | M
| 123
| 64.98±15.73
| 70
| -1.78
| 2.94
| 0.099
|
| -
| w/w*; 255B/+; p35/+ | F
| 123
| 74.41±5.98
| 76
| ---------
| ---------
| ---------
|
| L
| w/w*; 255B/+; p35/+ | F
| 0
| NA
| NA
| ---------
| ---------
| ---------
|
| L1-10
| w/w*; 255B/+; p35/+ | F
| 123
| 74.37±11.95
| 78
| -0.04
| 2.63
| 0.003
|
| Exp6 Life span assay of
two UAS-p35 lines with Elav driver at 25C |
| 8-5
| -
| yw/Y; +/+; Elav/+ | M
| 108
| 61.69±17.95
| 67
| ---------
| ---------
| ---------
|
| L1-10
| yw/Y; +/+; Elav/+ | M
| 115
| 54.17±15.55
| 58
| -12.18
| -13.43
| 1.80E-08
|
| -
| yw/w; +/+; Elav/+ | F
| 120
| 57.42±14.79
| 64
| ---------
| ---------
| ---------
|
| L1-10
| yw/w; +/+; Elav/+ | F
| 117
| 56.41±10.82
| 58
| -1.75
| -9.38
| 0.004
|
| 10-5
| -
| yw/Y; p35/+; Elav/+ | M
| 121
| 46.6±7.2
| 46
| ---------
| ---------
| ---------
|
| L1-10
| yw/Y; p35/+; Elav/+ | M
| 115
| 37.81±9.05
| 38
| -18.86
| -17.39
| 7.06E-14
|
| -
| yw/w*; p35/+; Elav/+ | F
| 123
| 50.63±15.32
| 54
| ---------
| ---------
| ---------
|
| L1-10
| yw/w*; p35/+; Elav/+ | F
| 121
| 49.19±12.16
| 50
| -2.85
| -7.41
| 0.035
|
| 11-5
| -
| yw/Y; +; Elav/p35 | M
| 120
| 54.32±13.04
| 56
| ---------
| ---------
| ---------
|
| L1-10
| yw/Y; +; Elav/p35 | M
| 111
| 52.31±10.98
| 52
| -3.7
| -7.14
| 0.037
|
| -
| yw/w*; +; Elav/p35 | F
| 123
| 52.7±13.97
| 54
| ---------
| ---------
| ---------
|
| L1-10
| yw/w*; +; Elav/p35 | F
| 118
| 56.63±10.73
| 58
| 7.45
| 7.41
| 0.091
|
Table 4. Parameters for Gompertz-Makeham model and likelihood ratio test results.
| Parameters | L | - | chi2 | df | p Value | chi2 | df | p Value |
| Females
| | | | | | | | | |
| | one parameter compared at
each time
| | | |
| p35 (X)
| | | | | | | Both a and b are constrained
|
| a
| 5.39 x 10-9 | 3.96 x 10-7 | 1.789
| 1
| 0.181
| | | |
| b
| 3.89 x 10-1 | 3.00 x 10-1 | 1.516
| 1
| 0.218
| | | |
| c
| 2.08 x 10-2 | 1.63 x 10-3 | 59.967
| 1
| <0.001 | 57.983
| 1
| <0.001 |
| | | | | | | | | |
| p35 (2)
| | | | | | | b is constrained
|
| a
| 7.71 x 10-6 | 2.10 x 10-4 | 5.234
| 1
| 0.022 | 50.203
| 1
| <0.001 |
| b
| 2.41 x 10-1 | 1.92 x 10-1 | 1.700
| 1
| 0.192
| | | |
| c
| 2.52 x 10-2 | 1.37 x 10-3 | 50.610
| 1
| <0.001 | 50.154
| 1
| <0.001 |
| | | | | | | | | |
| p35 (3)
| | | | | | | Both a and b are constrained
|
| a
| 3.31 x 10-6 | 3.00 x 10-6 | 0.003
| 1
| 0.958
| | | |
| b
| 2.50 x 10-1 | 2.46 x 10-1 | 0.009
| 1
| 0.923
| | | |
| c
| 1.36 x 10-2 | 1.80 x 10-4 | 46.090
| 1
| <0.001 | 66.787
| | <0.001 |