Long noncoding RNA LINC00958 accelerates the proliferation and matrix degradation of the nucleus pulposus by regulating miR-203/SMAD3
Abstract
Emerging evidence suggests that long noncoding RNAs (lncRNAs) play important roles in the development of intervertebral disc degeneration (IDD). LncRNA LINC00958 has recently been shown to play crucial roles in the development of tumors. However, the role of LINC00958 in IDD remains unclear. We showed that the expression of lncRNA LINC00958 was upregulated in degenerative NP samples, and LINC00958 expression increased gradually along with the grade of exacerbation of disc degeneration. Ectopic expression of LINC00958 promoted nucleus pulposus (NP) cell proliferation, inhibited aggrecan and Col II expression and promoted MMP-2 and MMP-13 expression. In addition, we showed that miR-203 expression was downregulated in degenerative NP samples, and miR-203 expression reduced gradually along with the grade of exacerbation of disc degeneration. Moreover, we demonstrated that the expression of miR-203 was inversely related with LINC00958 expression in NP samples. Ectopic expression of miR-203 inhibited NP cell growth and inhibited ECM degradation. Furthermore, we showed that ectopic expression of miR-203 suppressed the luciferase activity of the wild-type LINC00958 3'-UTR but not the mutant LINC00958 3'-UTR. Elevated expression of LINC00958 inhibited the expression of miR-203 and promoted the expression of SMAD3. In addition, we demonstrated that lncRNA LINC00958 exerted its function by targeting miR-203 in the NP cells. These data suggested that dysregulated lncRNA LINC00958 expression might play an important role in the development of IDD.
Introduction
Intervertebral disc degeneration (IDD) is one of the most common causes of low back pain (LBP) [1–4]. Approximately 80 percent of the population suffers from LBP at some point in their lifetime, while 10 percent of people with LBP become chronically disabled [5–7]. Although IDD is deemed to a natural process of intervertebral disc aging, several studies have demonstrated accelerated IDD due to genetic and environmental factors [1, 8–10]. IDD is characterized by the degradation of collagen, aggrecan and proteoglycans in the extracellular matrix (ECM) and nucleus pulposus (NP) cell proliferation, resulting in disrupting the homeostasis of NP and shifting intervertebral disc maintenance towards a catabolic and degenerative state [11–15]. Increasing evidence has shown that many cellular processes are involved in IDD [16–19]. However, the molecular process and mechanism of IDD remains unclear.
Long noncoding RNAs (lncRNAs) are a group of RNAs that are longer than 200 nts and that have no ability or limited ability to be coded into a protein [20–24]. Growing studies suggest that lncRNAs play crucial biological roles in diverse cellular processes including cell apoptosis, stem cell differentiation, proliferation and meiotic entry [25–29]. Moreover, emerging evidence has shown that lncRNAs were dysregulated in many tumors such as gastric cancer, hepatocellular carcinoma, osteosarcoma and lung cancer [30–33]. Recently, studies also found that lncRNAs played a critical role in the development of IDD [34–36]. For instance, Ruan et al. showed that the expression of lncRNA NEAT1 was upregulated in degenerated IVD tissues, and NEAT1 overexpression suppressed the expression of MMP13 and ADAMTS5 and induced collagen II and aggrecan expression, partly regulating the ERK/MAPK pathway. Mi et al. reported that FAF1 was overexpressed in IDD tissues, and ectopic expression of FAF1 induced NP cell growth by regulating the ERK signaling pathway. LINC00958 has recently been shown to play crucial roles in the development of tumors. For example, Seitz and colleagues first investigated the role of LINC00958 in bladder cancer [37]. They showed that LINC00958 was upregulated in bladder cancer, and knockdown of LINC00958 suppressed cell migration and viability. Guo et al. [38] reported that LINC00958 expression was upregulated in glioma cell lines and tissues, and knockdown of LINC00958 inhibited the invasion and proliferation of glioma cells by regulating miR-203 expression. Previous studies have reported that miR-203 increased the apoptosis and inflammation induced by lipopolysaccharide (LPS) by regulating NFIL3 in cardiomyocytes [39]. Given that LINC00958 and miR-203 are usually involved in the regulation of cell growth in several pathological processes and IDD is characterized by abnormal proliferation of NP cells, we supposed that LINC00958 may be overexpressed in IDD, thereby inducing NP cell growth. The major purpose of our study was to determine the role of LINC00958 in the development of IDD, the association between LINC00958 and miR-203 and their underlying mechanism in IDD.
Results
LncRNA LINC00958 was upregulated in degenerative NP samples
To study the role of lncRNA LINC00958 in IDD development, we first analyzed the expression of LINC00958 in the NP tissues and the scoliotic NP samples. As shown in Figure 1A, a notably upregulated level of LINC00958 was observed in NP samples with IDD compared with the expression levels in scoliotic tissues. Furthermore, we discovered that LINC00958 expression increased gradually along with the grade of exacerbation of disc degeneration (Figure 1B).
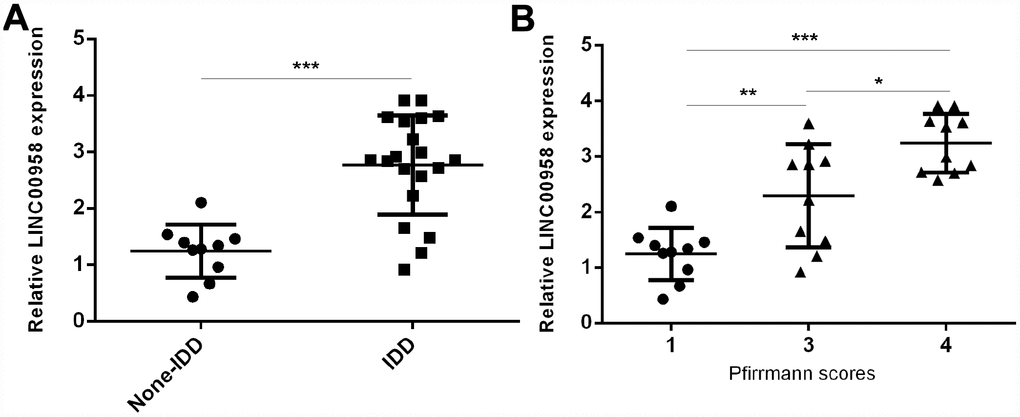
Figure 1. LncRNA LINC00958 was upregulated in degenerative NP samples. (A) The expression of LINC00958 was determined in 20 degenerative NP tissues and 10 scoliotic NP samples by using qRT-PCR analysis. (B) LINC00958 expression increased gradually along with the grade of exacerbation of disc degeneration. Data were showed as mean±SD. *p<0.05, **p<0.01 and ***p<0.001. U6 was used as the internal control.
miR-203 expression was downregulated in degenerative NP samples
Next, we investigated the expression of miR-203 in the NP tissues and the scoliotic NP samples. As shown in Figure 2A, a notably downregulated level of miR-203 was observed in NP samples with IDD compared with the expression level in scoliotic tissues. Furthermore, we showed that miR-203 expression decreased gradually along with the grade of exacerbation of disc degeneration (Figure 2B). Moreover, we demonstrated that the expression of miR-203 was inversely related with LINC00958 expression in NP samples (Figure 2C).
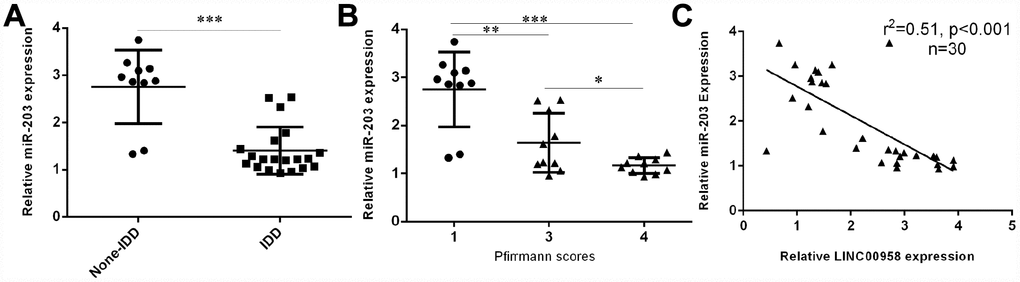
Figure 2. miR-203 expression was downregulated in degenerative NP samples. (A) The expression of miR-203 was determined in 20 degenerative NP tissues and 10 scoliotic NP samples by using qRT-PCR analysis. (B) miR-203 expression decreased gradually along with the grade of exacerbation of disc degeneration. (C) The expression of miR-203 was inversely related with LINC00958 expression in NP samples. *p<0.05, **p<0.01 and ***p<0.001. U6 was used as the internal control. Data were showed as mean±SD.
Interaction of lncRNA LINC00958 with miR-203
To investigate the mechanism underlying the effect of LINC00958 on NP cell function, bioinformatics analysis was performed to find a potential target gene of LINC00958. Using qRT-PCR, we showed that overexpression of miR-203 enhanced miR-203 expression in the NP cell (Figure 3A). In addition, a dual luciferase reporter analysis indicated that ectopic expression of miR-203 suppressed the luciferase activity of the wild-type LINC00958 3'-UTR but not the mutant LINC00958 3'-UTR (Figure 3B). Moreover, the expression of LINC00958 was significantly upregulated in the NP cell after it was transfected with pcDNA-LINC00958 (Figure 3C). Elevated expression of LINC00958 decreased the expression of miR-203 in the NP cells (Figure 3D). Overexpression of LINC00958 increased the mRNA and protein expression of SMAD3 in the NP cells (Figure 3E and 3F).
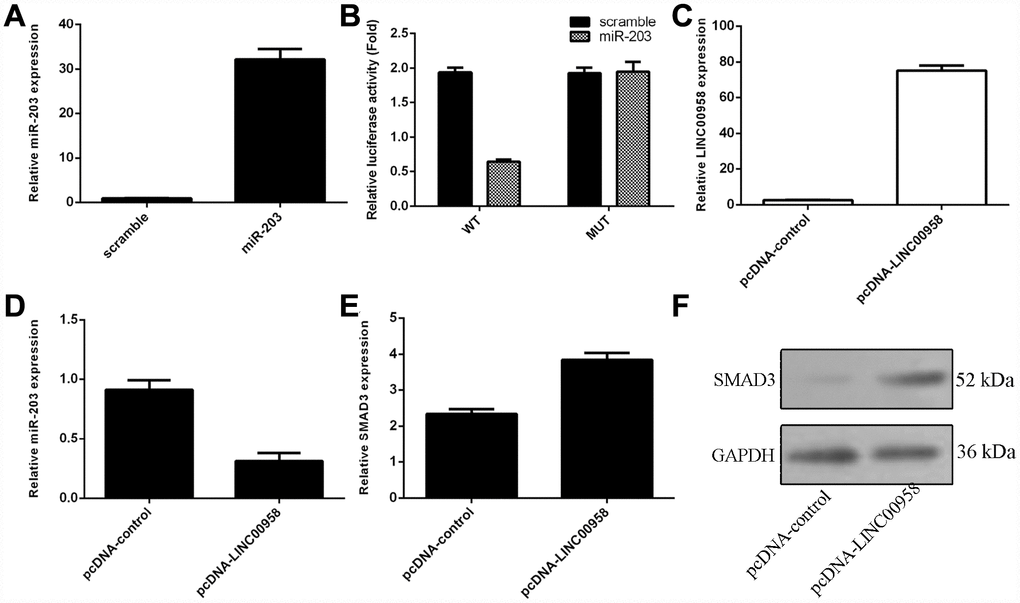
Figure 3. The interaction of lncRNA LINC00958 with miR-203. (A) The expression of miR-203 was measured in the NP cells by using qRT-PCR analysis. (B) Ectopic expression of miR-203 suppressed the luciferase activity of the wild-type LINC00958 3'-UTR but not the mutant LINC00958 3'-UTR. (C) The expression of LINC00958 was measured in the NP cells by using qRT-PCR analysis. (D) Elevated expression of LINC00958 decreased the expression of miR-203 in the NP cells. (E) Overexpression of LINC00958 promoted the mRNA expression of SMAD3 in the NP cells. (F) The protein expression of SMAD3 was detected by western blot, and GAPDH was used as the loading control. U6 was used as the internal control for miR-203. Data were showed as mean±SD.
Ectopic expression of LINC00958 promoted NP cell proliferation
Next, we studied the functional role of LINC00958 in the NP cells. Ectopic expression of LINC00958 promoted NP cell growth, which was determined by using the MTT assay (Figure 4A). In addition, we showed that overexpression of LINC00958 increased cyclin D1 expression in the NP cells (Figure 4B). Moreover, elevated LINC00958 expression enhanced PCNA expression in the NP cells (Figure 4C). Furthermore, we proved that LINC00958 overexpression promoted the protein expression of cyclin D1 and PCNA (Figure 4D).
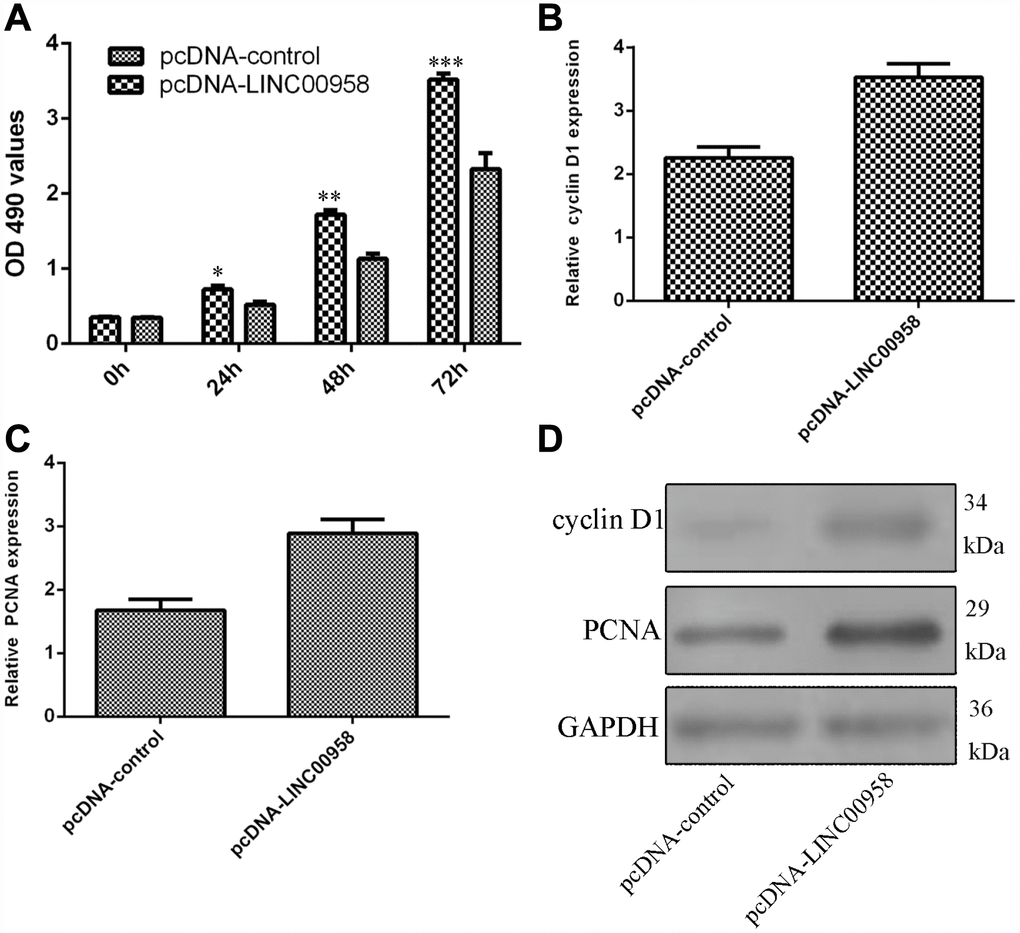
Figure 4. Ectopic expression of LINC00958 promoted NP cell proliferation. (A) Ectopic expression of LINC00958 promoted NP cell growth, which was determined by using the MTT assay. (B) Overexpression of LINC00958 increased cyclin D1 expression in the NP cell. (C) Elevated LINC00958 expression enhanced PCNA expression in the NP cell. (D) LINC00958 overexpression promoted the protein expression of cyclin D1 and PCNA. *p<0.05, **p<0.01 and ***p<0.001. GAPDH was used as the loading control. Data were showed as mean±SD.
Overexpression of LINC00958 inhibited aggrecan and Col II expression and promoted MMP-2 and MMP-13 expression
Elevated expression of LINC00958 increased MMP-2 expression in the NP cells (Figure 5A). In addition, we showed that LINC00958 overexpression promoted MMP-13 expression in the NP cells (Figure 5B). Moreover, we indicated that elevated expression of LINC00958 suppressed Col II expression in the NP cells (Figure 5C). Overexpression of LINC00958 inhibited aggrecan expression in the NP cells (Figure 5D).
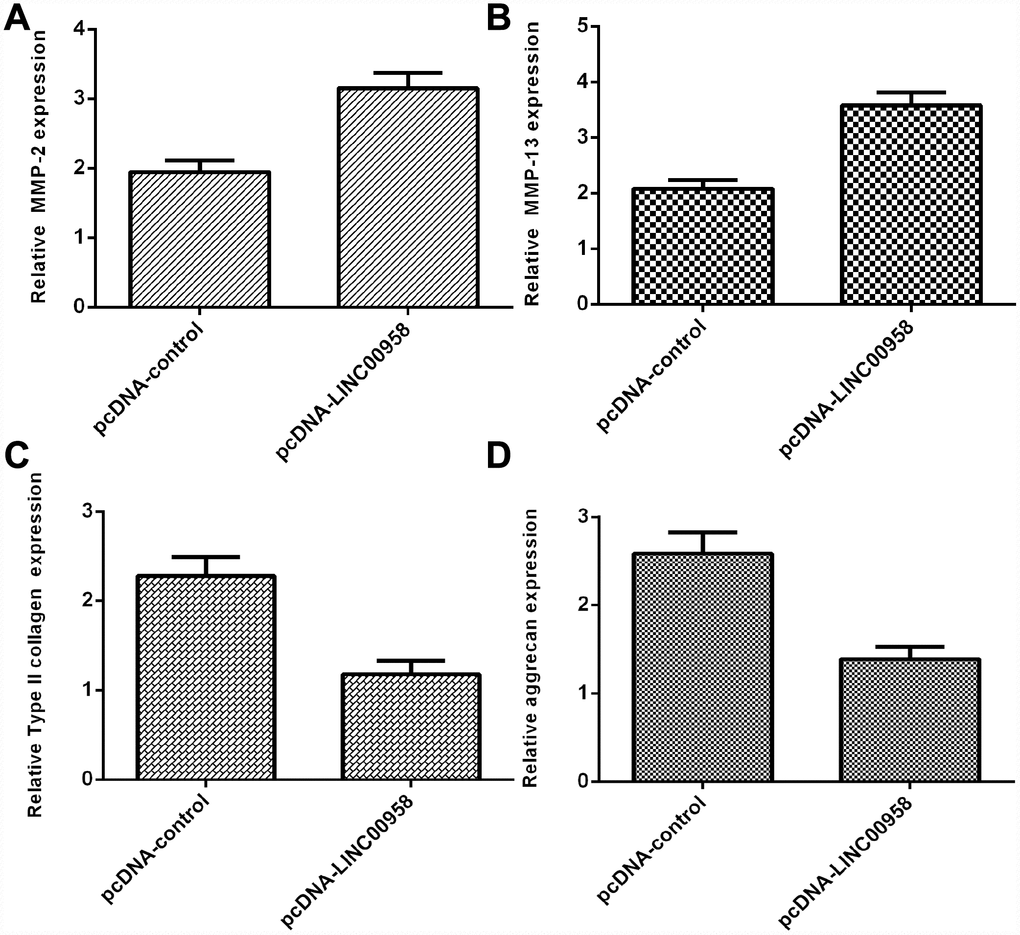
Figure 5. Overexpression of LINC00958 inhibited aggrecan and Col II expression and promoted MMP-2 and MMP-13 expression. (A) The expression of MMP-2 was determined by using qRT-PCR analysis. (B) The expression of MMP-2 was determined by using qRT-PCR analysis. (C) Elevated expression of LINC00958 suppressed Col II expression in the NP cells. (D) Overexpression of LINC00958 inhibited aggrecan expression in the NP cells. GAPDH was used as the internal control. Data were showed as mean±SD.
Ectopic expression of miR-203 decreased NP cell growth and inhibited ECM degradation
We discovered that elevated expression of miR-203 inhibited NP cell proliferation by using MTT analysis (Figure 6A). Ectopic miR-203 expression inhibited the expression of cyclin D1 in the NP cells (Figure 6B). Elevated expression of miR-203 suppressed PCNA expression in the NP cells (Figure 6C). In addition, we showed that overexpression of miR-203 suppressed the expression of MMP-2 and MMP-13 in the NP cells (Figure 6D and 6E). Moreover, elevated expression of miR-203 induced the expression of Col II and aggrecan in the NP cells (Figure 6F and 6G).
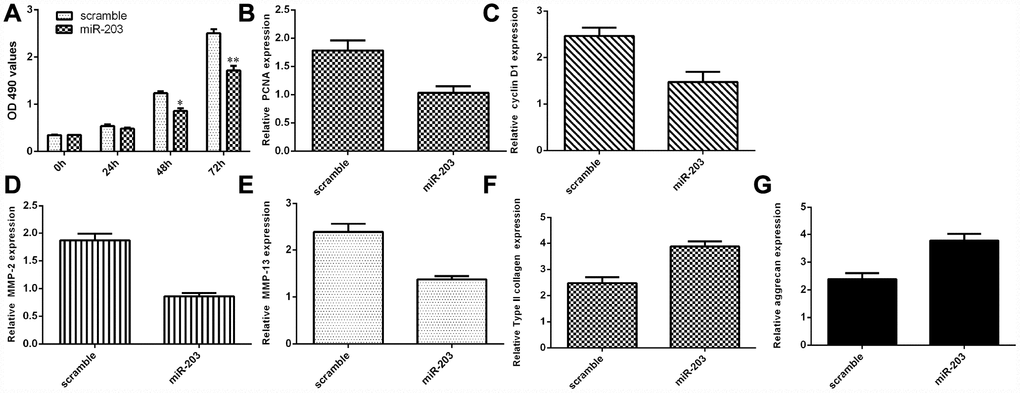
Figure 6. Ectopic expression of miR-203 decreased NP cell growth and inhibited ECM degradation. (A) Elevated expression of miR-203 inhibited NP cell proliferation, which was determined by MTT analysis. (B) The expression of PCNA was determined by using qRT-PCR analysis. (C) Elevated expression of miR-203 suppressed PCNA expression in the NP cells. (D) The expression of MMP-2 was determined by using qRT-PCR assay. (E) The expression of MMP-13 was determined by using qRT-PCR assay. (F) The expression of Col II was determined by using qRT-PCR assay. (G) The expression of aggrecan was determined by using qRT-PCR assay. *p<0.05, **p<0.01. GAPDH was used as the internal control. Data were showed as mean±SD.
LncRNA LINC00958 exerted its function by targeting miR-203 in the NP cells
To determine whether the functional effect of LINC00958 in promoting NP proliferation and ECM degradation was due to targeting miR-203, miR-203 was overexpressed in the NP cells through transfection with miR-203 mimic and pcDNA-LINC00958. The MTT analysis indicated that ectopic expression of miR-203 inhibited cell proliferation in the LINC00958-overexpressing NP cells (Figure 7A). In line with this result, the qRT-PCR assay showed that miR-203 overexpression suppressed cyclin D1 (Figure 7B) and PCNA (Figure 7C) expression in the LINC00958-overexpressing NP cells. Furthermore, the qRT-PCR assay demonstrated that elevated expression of LINC00958 suppressed MMP-3 (Figure 7D) and MMP-13 (Figure 7E) expression and enhanced Col II (Figure 7F) and aggrecan (Figure 7G) expression in the LINC00958-overexpressing NP cells.
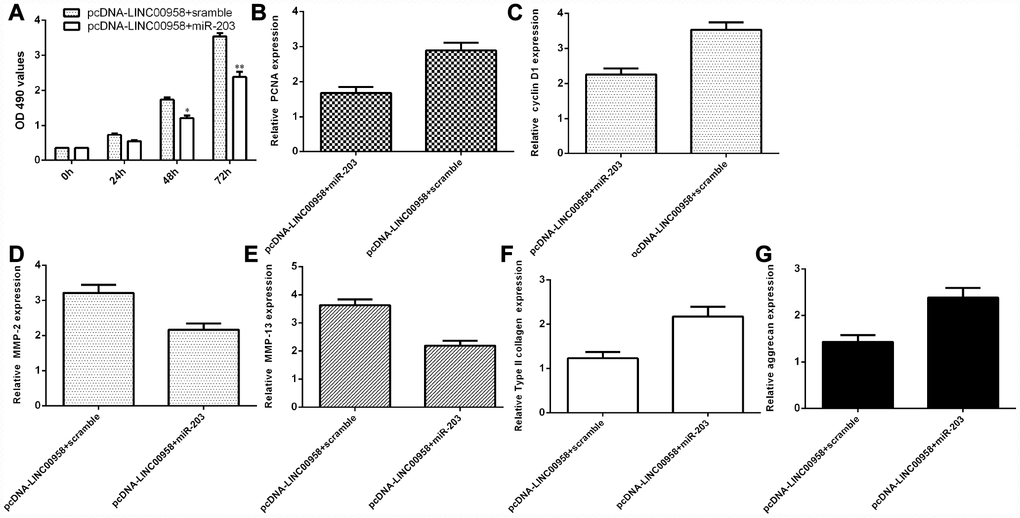
Figure 7. LncRNA LINC00958 exerted its function by targeting miR-203 in the NP cells. (A) Cell proliferation was measured by using the MTT assay. (B) PCNA expression was determined by qRT-PCR. (C) The expression of cyclin D1 was measured by qRT-PCR. (D) Elevated expression of LINC00958 suppressed the expression of MMP-3 in the LINC00958-overexpressing NP cells. (E) The expression of MMP-13 was determined by qRT-PCR analysis. (F) Col II expression was measured by using qRT-PCR analysis. (G) The expression of aggrecan was detected by using qRT-PCR analysis. *p<0.05, **p<0.01. GAPDH was used as the internal control. Data were showed as mean±SD.
Discussion
In our study, we showed that the expression of lncRNA LINC00958 was upregulated in degenerative NP samples, and LINC00958 expression increased gradually along with the grade of exacerbation of disc degeneration. Ectopic expression of LINC00958 promoted NP cell proliferation, inhibited aggrecan and Col II expression and promoted MMP-2 and MMP-13 expression. In addition, we showed that miR-203 expression was downregulated in degenerative NP samples, and miR-203 expression decreased gradually along with the grade of exacerbation of disc degeneration. Moreover, we demonstrated that the expression of miR-203 was inversely correlated with LINC00958 expression in NP samples. Ectopic expression of miR-203 inhibited NP cell growth and inhibited ECM degradation. Furthermore, we showed that ectopic expression of miR-203 suppressed the luciferase activity of the wild-type LINC00958 3'-UTR but not the mutant LINC00958 3'-UTR. Elevated expression of LINC00958 inhibited the expression of miR-203 and promoted the expression of SMAD3. In addition, we demonstrated that lncRNA LINC00958 exerted its function by targeting miR-203 in the NP cells. These data suggested that dysregulated lncRNA LINC00958 expression might play an important role in the development of IDD.
LINC00958 has been shown to play important roles in the development of tumors. For example, Seitz and colleagues investigated the role of LINC00958 in bladder cancer [37]. They used RNA-sequencing to find aberrantly expressed lncRNAs in bladder cancer. Their data suggested that LINC00958 was upregulated in bladder cancer and knockdown of LINC00958 suppressed cell migration and viability. Silencing of LINC00958 affected resistance to anoikis and invasion. Chen and colleagues showed that LINC00958 was overexpressed in endometrial cancer, and LINC01480 was shown to be required to express the micropeptide [40]. He and colleagues demonstrated that LINC00958 was upregulated in bladder tumor-associated lymphatic metastasis and lymphangiogenesis [41]. Ectopic expression of LINC00958 promoted VEGF-C expression by associating with WDR5. However, the expression and functional role of LINC00958 remained unknown. In our study, we first determined the expression of LINC00958 in the degenerative NP samples. We found that LINC00958 expression was upregulated in degenerative NP samples, and LINC00958 expression increased gradually along with the grade of exacerbation of disc degeneration. It is suggested that there is correlation between expression of LINC00958 and low back pain. In addition, we showed that LINC00958 overexpression induced NP cell proliferation, decreased aggrecan and Col II expression and promoted MMP-2 and MMP-13 expression.
Furthermore, we analyzed the potential mechanism of LINC00958 in the development of IDD. Previous studies have suggested that lncRNAs act as a molecular sponge of miRNA, modulating its expression [42, 43]. In addition, Guo et al. [38] reported that LINC00958 expression was upregulated in glioma cell lines and tissues, and knockdown of LINC00958 inhibited glioma cell invasion and proliferation by regulating miR-203 expression. Nevertheless, the relationship between LINC00958 and miR-203 in the function of NP cells remains to be studied. In our study, we demonstrated that ectopic expression of LINC00958 decreased the expression of miR-203 in NP cells. In addition, we demonstrated that miR-203 expression was downregulated in degenerative NP samples, and miR-203 expression decreased gradually along with the grade of exacerbation of disc degeneration. Moreover, we demonstrated that the expression of miR-203 was inversely related with LINC00958 expression in NP samples. Hu et al. [44] reported that miR-203 prevented the deposition and synthesis of ECM components via a SMAD3-dependent mechanism. In line with this finding, we demonstrated that ectopic expression of miR-203 inhibited NP cell growth and ECM degradation. Finally, we demonstrated that LncRNA LINC00958 exerted its function by targeting miR-203 in the NP cells.
Limitation
The major limitation of our research was that the control NP tissues are from scoliosis patients [45, 46]. Healthy age and spine level matched cadaveric discs were better to use as the control NP tissues. However, it is difficult to obtain the healthy age and spine level matched cadaveric discs.
In summary, we demonstrated that LINC00958 expression was upregulated in degenerative NP samples, and LINC00958 expression increased gradually along with the grade of exacerbation of disc degeneration. Ectopic expression of LINC00958 promoted NP cell proliferation, inhibited aggrecan and Col II expression and promoted MMP-2 and MMP-13 expression partly by regulating miR-203 expression. These results suggested that dysregulated LINC00958 expression may play an important role in the development of IDD.
Materials and Methods
Tissues
Human lumbar disc tissues were obtained from patients with IDD (n =20; average age 43 ± 7.4 years, range 31–56 years) or scoliosis (n= 10; average age 20 ± 1.2 years, range 18–22 years) who were undergoing spinal fusion at the China-Japan Union Hospital of Jilin University (Jilin, China). The tissues of the scoliosis patients were used as the control. These samples were immediately frozen in liquid nitrogen until they were used. The degree of disc degeneration was determined by a magnetic resonance imaging (MRI) scan following the modified Pfirrmann classification (Supplementary Table 1). This study was approved by the ethics committee of Jilin University. Written informed consent was collected from each patient.
Nucleus pulposus cell culture and transfection
Human nucleus pulposus (NP) cells were isolated and cultured following the procedures outlined in several previous studies [11–13]. NP cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 containing fetal bovine serum (FBS), streptomycin, penicillin and l-glutamine. pcDNA-control, pcDNA-LINC00958, miR-203 mimic and scramble were obtained from GenePharma (Shanghai, China). Cell transfection was performed by using Lipofectamine 3000 (Invitrogen, CA, USA) according to the manufacturer’s instructions.
RNA isolation and qRT-PCR
Total RNA was isolated from NP cells or samples with TRIzol (Invitrogen, CA, USA). The concentration of RNA was detected using a NanoDrop spectrophotometer (NanoDrop Technologies, DE, USA). cDNA (First-strand complementary DNA) synthesis was performed from 1 mg of RNA in a 12 ml volume containing dNTP Mix (Invitrogen, USA) and stem-loop primer. The mix was then incubated for 5 min at 65 °C and mixed with 0.1 M DTT, 5xRT buffer, RNase inhibitor and MultiScribe reverse transcriptase (Invitrogen, USA). Real-time quantitative polymerase chain reactions (qRT-PCRs) were performed to measure the expression of miRNA, lncRNA and mRNA using the SYBR Green PCR kit on the 7000 Sequence Detection System (Applied Biosystems, CA, USA). GAPDH and U6 were used as the internal controls for mRNA, lncRNA, and miRNA expression. The relative expression was determined by the 2−ΔΔCt method. The primer sequences used in this study were as follows: MMP-2, (forward) 5ʹ-GGCCCTGTCACTCCTGAG AT-3ʹ and (reverse) 5ʹ-GGCATCCAGGTTATCGGG GA-3ʹ; GADPH, (forward) 5ʹ-GGCCTCCAAGGAGT AAGACC-3ʹ and (reverse) 5ʹ-AGGGGTCTACAT GGCAACTG-3ʹ; SMAD3 (forward) 5ʹ-CGATGTCC CCAGCACACAATAAC-3ʹ and (reverse) 5ʹ-TAGTA GGAGATGGAGCACCAAAAGG-3ʹ; miR-203 (forward) 5ʹ- CGATGCTGTGAAATGTTTAGGGAC-3ʹ and (reverse) 5ʹ-TATGGTTTTGACGACTGTGT GAT-3ʹ; U6 (forward) 5ʹ-ATTGGAACGATACAG AGAAGATT-3ʹ and (reverse) 5ʹ-GGAACGCTTCACG AATTTG-3ʹ; and LINC00958, (forward) 5ʹ-CC ATTGAAGATACCACGCTGC-3ʹ and (reverse) 5ʹ-G GTTGTTGCCCAGGGTAGTG-3ʹ.
Western blotting
Total protein was isolated from NP cells or samples with RIPA buffer (Sigma, MO, USA). A total of 20 μg of protein was separated with 10% SDS-PAGE (sodium dodecyl sulphate-polyacrylamide) and transferred to membranes (Millipore, MA, USA). The membrane was incubated with the primary antibodies (SMAD3, catalogue: SAB4504210 and GAPDH, catalogue: G9545, Sigma; PCNA, catalogue: SAB2502098 and cyclin D1, catalogue: SAB1306586) at 4 °C overnight. Then, the membrane was labeled with horseradish peroxidase (HRP)-conjugated anti-IgG antibody. The protein signal was measured with the ECL system. GAPDH was used as the internal control.
Cell proliferation assays
Cell growth was evaluated by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) according to manufacturer’s information. Cells were cultured in a 96-well plate, and cell proliferation was measured at different time points. A total of 20 μl of MTT solution was added to each well, and they were cultured for an additional three hours. Then, the absorbance was determined at 490 nm using a Spectra microplate reader (Molecular Devices, CA, USA).
Luciferase reporter assay
Bioinformatics databases (http://starbase.sysu.edu.cn/index.php) were used to predict binding sequences between miR-203 and LINC00958. miR-203 was predicted to have putative binding sites of LINC00958. The seed sequences from LINC00958 and its mutant were subcloned into the downstream luciferase gene in the Dual-luciferase Expression Vector (Promega, WI). For the luciferase reporter analysis, cells were cultured in a 96-well plate and cotransfected with pmirGLO-LINC00958-wt or pmirGLO-LINC00958-mut and miR-203 mimic or scramble by the Lipofectamine 3000 reagent (Invitrogen, USA). The luciferase activity was detected by using the Dual-Luciferase Assay System (Promega).
Statistical analysis
Data are shown as the mean ± standard deviation (SD). Statistical analysis was performed using SPSS 18.0 (SPSS, IL, USA). The significant differences between groups were determined by analysis of variance or Student’s t test. A P value < 0.05 was considered statistically significant.
Conflicts of Interest
The authors declare that they have no conflicts of interests
Funding
This work was supported by the basic department of provincial science and technology, department of key laboratory of Jilin province for precision medicine of bone and joint degenerative diseases (No.20180622005JC).
References
-
1.
Liu W, Xia P, Feng J, Kang L, Huang M, Wang K, Song Y, Li S, Wu X, Yang S, Yang C. MicroRNA-132 upregulation promotes matrix degradation in intervertebral disc degeneration. Exp Cell Res. 2017; 359:39–49. https://doi.org/10.1016/j.yexcr.2017.08.011 [PubMed]
-
2.
Speed C. Low back pain. BMJ. 2004; 328:1119–21. https://doi.org/10.1136/bmj.328.7448.1119 [PubMed]
-
3.
Lee HJ, Seo JC, Kwak MA, Park SH, Min BM, Cho MS, Shin I, Jung JY, Roh WS. Acupuncture for low back pain due to spondylolisthesis: study protocol for a randomized controlled pilot trial. Trials. 2014; 15:105. https://doi.org/10.1186/1745-6215-15-105 [PubMed]
-
4.
Arana E, Kovacs FM, Royuela A, Estremera A, Asenjo B, Sarasíbar H, Amengual G, Galarraga I, Alonso A, Casillas C, Muriel A, Montoya J, Ordóñez C, et al. Modic changes and associated features in Southern European chronic low back pain patients. Spine J. 2011; 11:402–11. https://doi.org/10.1016/j.spinee.2011.03.019 [PubMed]
-
5.
Muraki S, Akune T, Oka H, En-Yo Y, Yoshida M, Saika A, Suzuki T, Yoshida H, Ishibashi H, Tokimura F, Yamamoto S, Nakamura K, Kawaguchi H, Yoshimura N. Health-related quality of life in subjects with low back pain and knee pain in a population-based cohort study of Japanese men: the Research on Osteoarthritis Against Disability study. Spine. 2011; 36:1312–19. https://doi.org/10.1097/BRS.0b013e3181fa60d1 [PubMed]
-
6.
Becker A, Held H, Redaelli M, Chenot JF, Leonhardt C, Keller S, Baum E, Pfingsten M, Hildebrandt J, Basler HD, Kochen MM, Donner-Banzhoff N, Strauch K. Implementation of a guideline for low back pain management in primary care: a cost-effectiveness analysis. Spine (Phila Pa 1976). 2012; 37:701–10. https://doi.org/10.1097/BRS.0b013e31822b01bd [PubMed]
-
7.
Millecamps M, Tajerian M, Naso L, Sage EH, Stone LS. Lumbar intervertebral disc degeneration associated with axial and radiating low back pain in ageing SPARC-null mice. Pain. 2012; 153:1167–79. https://doi.org/10.1016/j.pain.2012.01.027 [PubMed]
-
8.
Ye D, Dai L, Yao Y, Qin S, Xie H, Wang W, Liang W. miR-155 Inhibits Nucleus Pulposus Cells’ Degeneration through Targeting ERK ½. Dis Markers. 2016; 2016:6984270. https://doi.org/10.1155/2016/6984270 [PubMed]
-
9.
Gu SX, Li X, Hamilton JL, Chee A, Kc R, Chen D, An HS, Kim JS, Oh CD, Ma YZ, van Wijnen AJ, Im HJ. MicroRNA-146a reduces IL-1 dependent inflammatory responses in the intervertebral disc. Gene. 2015; 555:80–87. https://doi.org/10.1016/j.gene.2014.10.024 [PubMed]
-
10.
Liu H, Huang X, Liu X, Xiao S, Zhang Y, Xiang T, Shen X, Wang G, Sheng B. miR-21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int J Mol Sci. 2014; 15:4007–18. https://doi.org/10.3390/ijms15034007 [PubMed]
-
11.
Li Z, Liang J, Wu WK, Yu X, Yu J, Weng X, Shen J. Leptin activates RhoA/ROCK pathway to induce cytoskeleton remodeling in nucleus pulposus cells. Int J Mol Sci. 2014; 15:1176–88. https://doi.org/10.3390/ijms15011176 [PubMed]
-
12.
Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G, Liu J. Leptin induces cyclin D1 expression and proliferation of human nucleus pulposus cells via JAK/STAT, PI3K/Akt and MEK/ERK pathways. PLoS One. 2012; 7:e53176. https://doi.org/10.1371/journal.pone.0053176 [PubMed]
-
13.
Li Z, Shen J, Wu WK, Yu X, Liang J, Qiu G, Liu J. The role of leptin on the organization and expression of cytoskeleton elements in nucleus pulposus cells. J Orthop Res. 2013; 31:847–57. https://doi.org/10.1002/jor.22308 [PubMed]
-
14.
Lv F, Huang Y, Lv W, Yang L, Li F, Fan J, Sun J. MicroRNA-146a Ameliorates Inflammation via TRAF6/NF-κB Pathway in Intervertebral Disc Cells. Med Sci Monit. 2017; 23:659–64. https://doi.org/10.12659/MSM.898660 [PubMed]
-
15.
Studer RK, Vo N, Sowa G, Ondeck C, Kang J. Human nucleus pulposus cells react to IL-6: independent actions and amplification of response to IL-1 and TNF-α. Spine. 2011; 36:593–99. https://doi.org/10.1097/BRS.0b013e3181da38d5 [PubMed]
-
16.
Tao B, Yi J, Huang C, Xu W, Qin C, Chen L, Chen J, Gao Y, Wang R. microRNA-96 regulates the proliferation of nucleus pulposus cells by targeting ARID2/AKT signaling. Mol Med Rep. 2017; 16:7553–60. https://doi.org/10.3892/mmr.2017.7560 [PubMed]
-
17.
Wang T, Li P, Ma X, Tian P, Han C, Zang J, Kong J, Yan H. MicroRNA-494 inhibition protects nucleus pulposus cells from TNF-α-induced apoptosis by targeting JunD. Biochimie. 2015; 115:1–7. https://doi.org/10.1016/j.biochi.2015.04.011 [PubMed]
-
18.
Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X, Qiu G. MicroRNA-10b promotes nucleus pulposus cell proliferation through RhoC-Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PLoS One. 2013; 8:e83080. https://doi.org/10.1371/journal.pone.0083080 [PubMed]
-
19.
Zhao K, Zhang Y, Kang L, Song Y, Wang K, Li S, Wu X, Hua W, Shao Z, Yang S, Yang C. Epigenetic silencing of miRNA-143 regulates apoptosis by targeting BCL2 in human intervertebral disc degeneration. Gene. 2017; 628:259–66. https://doi.org/10.1016/j.gene.2017.07.043 [PubMed]
-
20.
McCleland ML, Mesh K, Lorenzana E, Chopra VS, Segal E, Watanabe C, Haley B, Mayba O, Yaylaoglu M, Gnad F, Firestein R. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Invest. 2016; 126:639–52. https://doi.org/10.1172/JCI83265 [PubMed]
-
21.
Chen ZH, Hu HK, Zhang CR, Lu CY, Bao Y, Cai Z, Zou YX, Hu GH, Jiang L. Down-regulation of long non-coding RNA FOXD3 antisense RNA 1 (FOXD3-AS1) inhibits cell proliferation, migration, and invasion in malignant glioma cells. Am J Transl Res. 2016; 8:4106–19. [PubMed]
-
22.
Dou J, Ni Y, He X, Wu D, Li M, Wu S, Zhang R, Guo M, Zhao F. Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am J Transl Res. 2016; 8:98–108. [PubMed]
-
23.
Yang C, Wu K, Wang S, Wei G. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem. 2018; 119:5646–56. https://doi.org/10.1002/jcb.26743 [PubMed]
-
24.
Xin Y, Li Z, Shen J, Chan MT, Wu WK. CCAT1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016; 49:255–60. https://doi.org/10.1111/cpr.12252 [PubMed]
-
25.
Zhao J, Zhang C, Gao Z, Wu H, Gu R, Jiang R. Long non-coding RNA ASBEL promotes osteosarcoma cell proliferation, migration, and invasion by regulating microRNA-21. J Cell Biochem. 2018; 119:6461–69. https://doi.org/10.1002/jcb.26671 [PubMed]
-
26.
Zhang XY, Tang XY, Ma LJ, Guo YL, Li XS, Zhao LM, Tian CJ, Cheng DJ, Chen ZC, Zhang LX. Schisandrin B down-regulated lncRNA BCYRN1 expression of airway smooth muscle cells by improving miR-150 expression to inhibit the proliferation and migration of ASMC in asthmatic rats. Cell Prolif. 2017; 50:e12382. https://doi.org/10.1111/cpr.12382 [PubMed]
-
27.
Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017; 50:e12389. https://doi.org/10.1111/cpr.12389 [PubMed]
-
28.
Li Z, Shen J, Chan MT, Wu WK. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016; 49:471–75. https://doi.org/10.1111/cpr.12269 [PubMed]
-
29.
Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y, Jiang X. LncRNA-ATB: an indispensable cancer-related long noncoding RNA. Cell Prolif. 2017; 50:e12381. https://doi.org/10.1111/cpr.12381 [PubMed]
-
30.
She K, Yan H, Huang J, Zhou H, He J. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. 2018; 51. https://doi.org/10.1111/cpr.12406 [PubMed]
-
31.
Zhang Y, Yang R, Lian J, Xu H. LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res. 2016; 8:5035–43. [PubMed]
-
32.
Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res. 2016; 8:4095–105. [PubMed]
-
33.
Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, Wang P, Sun B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol Carcinog. 2016; 55:209–19. https://doi.org/10.1002/mc.22270 [PubMed]
-
34.
Chen WK, Yu XH, Yang W, Wang C, He WS, Yan YG, Zhang J, Wang WJ. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017; 50. https://doi.org/10.1111/cpr.12313 [PubMed]
-
35.
Ruan Z, Ma H, Li J, Liu H, Jia H, Li F. The long non-coding RNA NEAT1 contributes to extracellular matrix degradation in degenerative human nucleus pulposus cells. Exp Biol Med (Maywood). 2018; 243:595–600. https://doi.org/10.1177/1535370218760774 [PubMed]
-
36.
Wang X, Lv G, Li J, Wang B, Zhang Q, Lu C. LncRNA-RP11-296A18.3/miR-138/HIF1A Pathway Regulates the Proliferation ECM Synthesis of Human Nucleus Pulposus Cells (HNPCs). J Cell Biochem. 2017; 118:4862–71. https://doi.org/10.1002/jcb.26166 [PubMed]
-
37.
Seitz AK, Christensen LL, Christensen E, Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM, Palmfeldt J, Thomson M, Jensen MT, Nawroth R, Maurer T, et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci Rep. 2017; 7:395. https://doi.org/10.1038/s41598-017-00327-0 [PubMed]
-
38.
Guo E, Liang C, He X, Song G, Liu H, Lv Z, Guan J, Yang D, Zheng J. Long Noncoding RNA LINC00958 Accelerates Gliomagenesis Through Regulating miR-203/CDK2. DNA Cell Biol. 2018; 37:465–72. https://doi.org/10.1089/dna.2018.4163 [PubMed]
-
39.
Li Y, Liu X, Du A, Zhu X, Yu B. miR-203 accelerates apoptosis and inflammation induced by LPS via targeting NFIL3 in cardiomyocytes. J Cell Biochem. 2019; 120:6605–6613. https://doi.org/10.1002/jcb.27955 [PubMed]
-
40.
Chen BJ, Byrne FL, Takenaka K, Modesitt SC, Olzomer EM, Mills JD, Farrell R, Hoehn KL, Janitz M. Transcriptome landscape of long intergenic non-coding RNAs in endometrial cancer. Gynecol Oncol. 2017; 147:654–62. https://doi.org/10.1016/j.ygyno.2017.10.006 [PubMed]
-
41.
He W, Zhong G, Jiang N, Wang B, Fan X, Chen C, Chen X, Huang J, Lin T. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest. 2018; 128:861–75. https://doi.org/10.1172/JCI96218 [PubMed]
-
42.
Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin-induced cytotoxicity via regulating microRNA let-7a in nasopharyngeal carcinoma. J Biochem Mol Toxicol. 2017; 31:e21904. https://doi.org/10.1002/jbt.21904 [PubMed]
-
43.
Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014; 5:2276–92. https://doi.org/10.18632/oncotarget.1902 [PubMed]
-
44.
Hu D, Hu Y, Xu W, Yu H, Yang N, Ni S, Fu R. miR-203 inhibits the expression of collagen-related genes and the proliferation of hepatic stellate cells through a SMAD3-dependent mechanism. Mol Med Rep. 2017; 16:1248–54. https://doi.org/10.3892/mmr.2017.6702 [PubMed]
-
45.
Walter BA, Korecki CL, Purmessur D, Roughley PJ, Michalek AJ, Iatridis JC. Complex loading affects intervertebral disc mechanics and biology. Osteoarthritis Cartilage. 2011; 19:1011–18. https://doi.org/10.1016/j.joca.2011.04.005 [PubMed]
-
46.
McDonnell P, Harrison N, McHugh PE. Investigation of the failure behaviour of vertebral trabecular architectures under uni-axial compression and wedge action loading conditions. Med Eng Phys. 2010; 32:569–76. https://doi.org/10.1016/j.medengphy.2010.02.005 [PubMed]